2C46
 
 | | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | | Descriptor: | MRNA CAPPING ENZYME | | Authors: | Debreczeni, J, Johansson, C, Longman, E, Gileadi, O, SavitskySmee, P, Smee, C, Bunkoczi, G, Ugochukwu, E, von Delft, F, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human RNA Guanylyltransferase and 5'-Phosphatase
To be Published
|
|
1ZO3
 
 | | The P-site and P/E-site tRNA structures fitted to P/I site codon. | | Descriptor: | tRNA | | Authors: | Allen, G.S, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | The Cryo-EM Structure of a Translation Initiation Complex from Escherichia coli.
Cell(Cambridge,Mass.), 121, 2005
|
|
8SAM
 
 | |
8SAP
 
 | |
8SAO
 
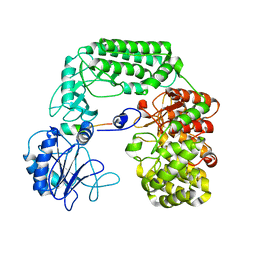 | |
7AEX
 
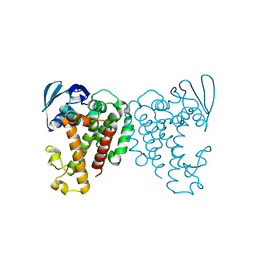 | |
3WFO
 
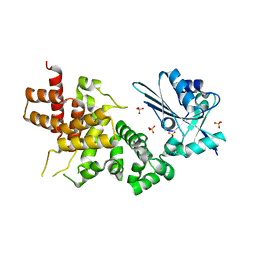 | | tRNA processing enzyme (apo form 1) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
2CQ4
 
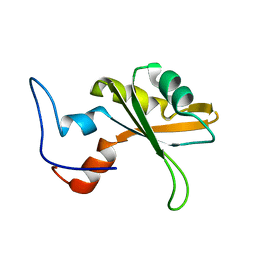 | | solution structure of RNA binding domain in RNA binding motif protein 23 | | Descriptor: | RNA binding motif protein 23 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RNA binding domain in RNA binding motif protein 23
To be Published
|
|
1O4X
 
 | |
4ILU
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | RNA polymerase-binding transcription factor CarD, SULFATE ION | | Authors: | Thakur, K.G, Kaur, G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
3WFP
 
 | | tRNA processing enzyme (apo form 2) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
5EGR
 
 | | tRNA guanine transglycosylase (TGT) in complex with an Immucillin derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-7-[(2~{S},3~{R},5~{S})-5-(hydroxymethyl)-3-oxidanyl-pyrrolidin-2-yl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of an Immucillin Derivative as a Class of tRNA-Guanine Transglycosylase Inhibitors: Exploration of a Transition State Analogous Binding Mode
To be Published
|
|
5HBY
 
 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue-3 binding sites | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*C)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Wang, J, Szostak, J.W. | | Deposit date: | 2016-01-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
2DIV
 
 | | Solution structure of the RRM domain of TRNA selenocysteine associated protein | | Descriptor: | TRNA selenocysteine associated protein | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of TRNA selenocysteine associated protein
To be Published
|
|
2KDV
 
 | |
6K84
 
 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
1HKN
 
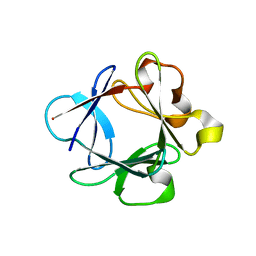 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
2K9L
 
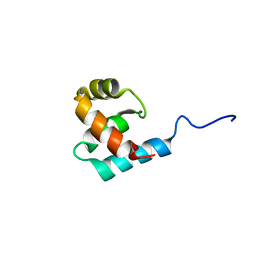 | | Structure of the Core Binding Domain of sigma54 | | Descriptor: | RNA polymerase sigma factor RpoN | | Authors: | Hong, E, Wemmer, D. | | Deposit date: | 2008-10-19 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA polymerase core-binding domain of sigma(54) reveals a likely conformational fracture point
J.Mol.Biol., 390, 2009
|
|
5MRX
 
 | |
1WEL
 
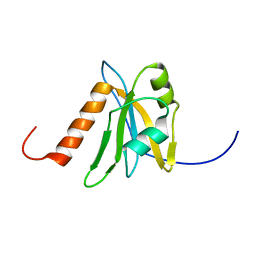 | | Solution structure of RNA binding domain in NP_006038 | | Descriptor: | RNA-binding protein 12 | | Authors: | Someya, T, Muto, Y, Nagata, T, Suzuki, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in NP_006038
To be Published
|
|
359D
 
 | |
1L8B
 
 | | Cocrystal Structure of the Messenger RNA 5' Cap-binding Protein (eIF4E) bound to 7-methylGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Niedzwiecka, A, Marcotrigiano, J, Stepinski, J, Jankowska-Anyszka, M, Wyslouch-Cieszynska, A, Dadlez, M, Gingras, A.-C, Mak, P, Darzynkiewicz, E, Sonenberg, N. | | Deposit date: | 2002-03-19 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5' cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins.
J.Mol.Biol., 319, 2002
|
|
5DEK
 
 | | RNA octamer containing dT | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing dT | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
6ZOT
 
 | |
5D8T
 
 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
