5NMR
 
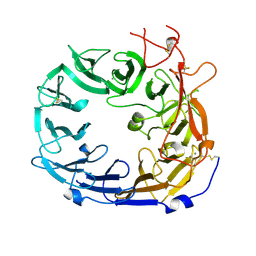 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
2NMR
 
 | |
2M4M
 
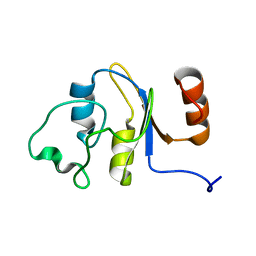 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2K27
 
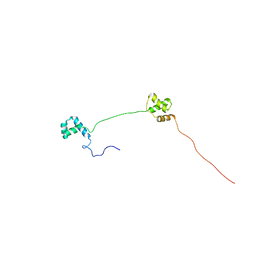 | | Solution structure of Human Pax8 Paired Box Domain | | Descriptor: | Paired box protein Pax-8 | | Authors: | Codutti, L, Esposito, G, Corazza, A, Fogolari, F, Tell, G, Vascotto, C, van Ingen, H, Boelens, R, Viglino, P, Quadrifoglio, F. | | Deposit date: | 2008-03-26 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of DNA-free Pax-8 Paired Box Domain Accounts for Redox Regulation of Transcriptional Activity in the Pax Protein Family.
J.Biol.Chem., 283, 2008
|
|
2ACF
 
 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab | | Authors: | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
2LAG
 
 | | Structure of the 44 kDa complex of interferon-alpha2 with the extracellular part of IFNAR2 obtained by 2D-double difference NOESY | | Descriptor: | Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Nudelman, I, Akabayov, S.R, Scherf, T, Anglister, J. | | Deposit date: | 2011-03-13 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
J.Am.Chem.Soc., 133, 2011
|
|
2P37
 
 | | Crystal structure of a lectin from Canavalia maritima seeds (CML) in complex with man1-3man-OMe | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Bezerra, G.A, Oliveira, T.M, Souza, E.P, Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W. | | Deposit date: | 2007-03-08 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
2P34
 
 | | Crystal structure of a lectin from Canavalia maritima seeds (CML) in complex with man1-4man-OMe | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Bezerra, G.A, Oliveira, T.M, Souza, E.P, Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W. | | Deposit date: | 2007-03-08 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: new insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
2OW4
 
 | | Crystal structure of a lectin from Canavalia maritima seeds (ConM) in complex with man1-2man-OMe | | Descriptor: | CALCIUM ION, Canavalia maritima lectin, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Bezerra, G.A, de Oliveira, T.M, de Souza, E.M, da Rocha, B.A.M, Benevides, R.G, Delatorre, P, Cavada, B.S, de Azevedo Jr, W.F. | | Deposit date: | 2007-02-15 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure-biological activity relationship in legume lectins.
J.Struct.Biol., 160, 2007
|
|
2CWM
 
 | | Native Crystal Structure of NO releasing inductive lectin from seeds of the Canavalia maritima (ConM) | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin | | Authors: | Cavada, B.S, De Azevedo Jr, W.F, Assreuy, A.M.S, Criddle, D.N, Gadelha, C.A.A, Delatorre, P, Souza, E.P, Rocha, B.A.M, Santi-Gadelha, T, Moreno, F.B.M.B. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native crystal structure of a nitric oxide-releasing lectin from the seeds of Canavalia maritima
J.Struct.Biol., 152, 2005
|
|
8SG2
 
 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
3H7W
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 | | Descriptor: | 2-nitro-N-(thiophen-3-ylmethyl)-4-(trifluoromethyl)aniline, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
8F4V
 
 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | Authors: | Bondarenko, V, Chen, Q, Tang, P. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
3H82
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(furan-2-ylmethyl)-2-nitro-4-(trifluoromethyl)aniline | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
5VJ8
 
 | |
2Y3L
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 2 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FBS
 
 | | Structure of monomeric NT from Euprosthenops australis Major Ampullate Spidroin 1 (MaSp1) | | Descriptor: | BROMIDE ION, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, M, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain
J.Mol.Biol., 2012
|
|
3TRX
 
 | |
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4UTT
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
5VMV
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with its double CpG-methylated DNA consensus binding site | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*TP*TP*CP*TP*(5CM)P*GP*(5CM)P*GP*AP*GP*AP*AP*GP*CP*A)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
5VMX
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
