3SZ8
 
 | |
2PLX
 
 | |
4DZ9
 
 | | hCA II in complex with novel sulfonamide inhibitors Set D | | Descriptor: | 4-[4-(cyclohexylmethyl)-1H-1,2,3-triazol-1-yl]-2,3,5,6-tetrafluorobenzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | hCA II in complex with novel sulfonamide inhibitors Set D
To be Published
|
|
2PQ8
 
 | | MYST histone acetyltransferase 1 | | Descriptor: | COENZYME A, Probable histone acetyltransferase MYST1, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MYST histone acetyltransferase 1.
To be Published
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
3TF2
 
 | | Crystal structure of the cap free human translation initiation factor eIF4E | | Descriptor: | Eukaryotic translation initiation factor 4E, UNKNOWN ATOM OR ION | | Authors: | Siddiqui, N, Tempel, W, Nedyalkova, L, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Borden, K.L.B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-15 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E.
J.Mol.Biol., 415, 2012
|
|
4E0M
 
 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway
to toxic amyloid aggregates
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2POE
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | Descriptor: | Cyclophilin-like protein, putative, FORMIC ACID | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
4H9Z
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Mn2+ | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Phosphotriesterase | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4HAE
 
 | | Crystal structure of the CDYL2-chromodomain | | Descriptor: | Chromodomain Y-like protein 2, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Dombrovski, L, Tempel, W, Dong, A, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the CDYL2-chromodomain
To be Published
|
|
4DWG
 
 | | Crystal structure of Trypanosome cruzi farnesyl diphosphate synthase in complex with [2-(n-heptylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4E3I
 
 | |
3G3Y
 
 | | Mth0212 in complex with ssDNA in space group P32 | | Descriptor: | 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-03 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
4E3W
 
 | |
3T4V
 
 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(2-napthalenemethyl)-L-cysteine | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
2O4T
 
 | |
3G5G
 
 | | Crystal Structure of the Wild-Type Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4DYK
 
 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
3TDS
 
 | | Crystal structure of HSC F194I | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3G8C
 
 | | Crystal Structure of Biotin Carboxylase in Complex with Biotin, Bicarbonate, ADP and Mg Ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, BIOTIN, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
4E0D
 
 | | Binary complex of Bacillus DNA Polymerase I Large Fragment E658A and duplex DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
2OA0
 
 | | Crystal structure of Calcium ATPase with bound ADP and cyclopiazonic acid | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Young, H.S, Moncoq, K.A. | | Deposit date: | 2006-12-14 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The molecular basis for cyclopiazonic Acid inhibition of the sarcoplasmic reticulum calcium pump.
J.Biol.Chem., 282, 2007
|
|
2OD7
 
 | | Crystal Structure of yHst2 bound to the intermediate analogue ADP-HPD, and and aceylated H4 peptide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Acetylated histone H4 peptide, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R.Q, Sanders, B.D. | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
4DZU
 
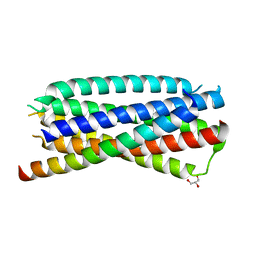 | | Complex of 3-alpha bound to gp41-5 | | Descriptor: | 3-alpha, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
3T8S
 
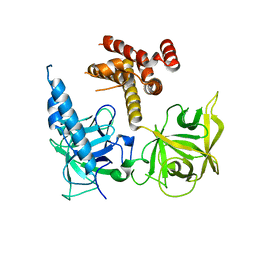 | | Apo and InsP3-bound Crystal Structures of the Ligand-Binding Domain of an InsP3 Receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Lin, C, Baek, K, Lu, Z. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Apo and InsP(3)-bound crystal structures of the ligand-binding domain of an InsP(3) receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
