4N08
 
 | |
1N1S
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
2OCX
 
 | | Crystal structure of Se-Met fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-12-21 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
4TRF
 
 | |
4TRU
 
 | |
3RJ8
 
 | | Crystal structure of carbohydrate oxidase from Microdochium nivale | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbohydrate oxidase, ... | | Authors: | Duskova, J, Skalova, T, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Kolenko, P, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
4DVH
 
 | | Crystal structure of Trypanosoma cruzi mitochondrial iron superoxide dismutase | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Larrieux, N, Buschiazzo, A. | | Deposit date: | 2012-02-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Molecular Basis of the Peroxynitrite-mediated Nitration and Inactivation of Trypanosoma cruzi Iron-Superoxide Dismutases (Fe-SODs) A and B: DISPARATE SUSCEPTIBILITIES DUE TO THE REPAIR OF TYR35 RADICAL BY CYS83 IN Fe-SODB THROUGH INTRAMOLECULAR ELECTRON TRANSFER.
J.Biol.Chem., 289, 2014
|
|
4CYK
 
 | | Structural basis for binding of Pan3 to Pan2 and its function in mRNA recruitment and deadenylation | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3, ZINC ION | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation.
Embo J., 33, 2014
|
|
5EQC
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
1I5G
 
 | | TRYPAREDOXIN II COMPLEXED WITH GLUTATHIONYLSPERMIDINE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONYLSPERMIDINE, TRYPAREDOXIN II | | Authors: | Hofmann, B, Budde, H, Bruns, K, Guerrero, S.A, Kalisz, H.M, Menge, U, Montemartini, M, Nogoceke, E, Steinert, P, Wissing, J.B, Flohe, L, Hecht, H.-J. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
6UVJ
 
 | |
6UWX
 
 | |
6N9P
 
 | | Discovery of affinity-based probes for Btk occupancy assay | | Descriptor: | N-(3-{[2-amino-3-(4-phenoxyphenyl)pyridin-4-yl]oxy}phenyl)propanamide, Tyrosine-protein kinase BTK | | Authors: | Mochalkin, I. | | Deposit date: | 2018-12-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery of Affinity-Based Probes for Btk Occupancy Assays.
ChemMedChem, 14, 2019
|
|
1YS3
 
 | |
1VZT
 
 | | ROLES OF INDIVIDUAL RESIDUES OF ALPHA-1,3 GALACTOSYLTRANSFERASES IN SUBSTRATE BINDING AND CATALYSIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, X, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-26 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
8B3E
 
 | | Variant Surface Glycoprotein VSG397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 397 | | Authors: | Zeelen, J.P, Stebbins, C.E, Dakovic, S, Foti, K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
5BSJ
 
 | |
5BS9
 
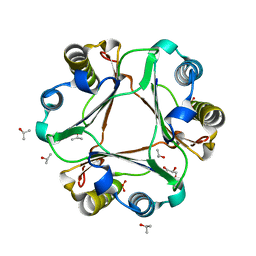 | |
4FER
 
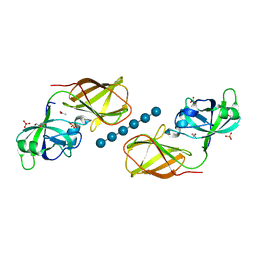 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose | | Descriptor: | ACETIC ACID, Expansin-yoaJ, GLYCEROL, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WJ8
 
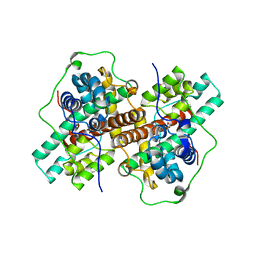 | | Crystal Structure of DL-2-haloacid dehalogenase mutant with 2-bromo-2-methylpropionate | | Descriptor: | 2-bromo-2-methylpropanoic acid, DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
1RDP
 
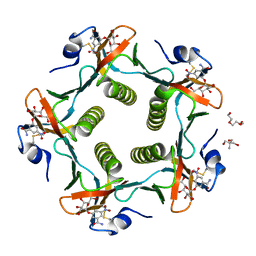 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RF2
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RD9
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1L0Z
 
 | | THE STRUCTURE OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH XENON AND BROMIDE, CRYOPROTECTED WITH DRY PARAFFIN OIL | | Descriptor: | BROMIDE ION, ELASTASE 1, SODIUM ION, ... | | Authors: | Tucker, P.A, Panjikar, S. | | Deposit date: | 2002-02-14 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6BAI
 
 | | Multiconformer model of apo K197C PTP1B at 100 K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
