8B9H
 
 | | Crystal structure of JAK2 JH2 in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B9E
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A3 | | Descriptor: | 6-[[methyl-[(3-methylthiophen-2-yl)methyl]amino]methyl]-~{N}4-phenyl-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8BAK
 
 | |
8BAB
 
 | | Crystal structure of JAK2 JH2-V617F in complex with CB76 | | Descriptor: | 6-[(1-methylimidazol-2-yl)sulfanylmethyl]-~{N}4-(3-methylphenyl)-1,3,5-triazine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B8N
 
 | |
8BA2
 
 | | Crystal structure of JAK2 JH2-V617F in complex with Z902-A1 | | Descriptor: | 6-[[(5-bromanylthiophen-2-yl)methyl-methyl-amino]methyl]-~{N}4-(4-methylphenyl)-1,3,5-triazine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B99
 
 | | Crystal structure of JAK2 JH2-V617F in complex with JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain.
Pharmaceuticals, 16, 2023
|
|
8B5L
 
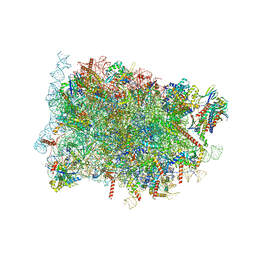 | | Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
8B6C
 
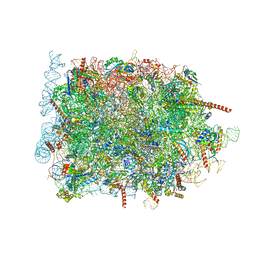 | | Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147 | | Descriptor: | 28S rRNA, 4-[[9-(cyclohexylmethyl)-3-methylidene-5-(4-methylphenyl)sulfonyl-1,5,9-triazacyclododec-1-yl]sulfonyl]-~{N},~{N}-dimethyl-aniline, 5.8S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
6UY8
 
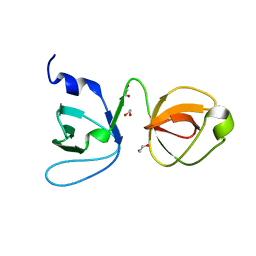 | |
8B7F
 
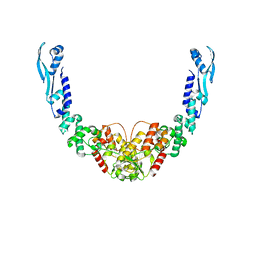 | | Nuclease from C. glutamicum | | Descriptor: | Ubiquitin-like protein SMT3,MksG | | Authors: | Wehenkel, A, Ben Assaya, M, Haouz, A. | | Deposit date: | 2022-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | The MksG nuclease is the executing part of the bacterial plasmid defense system MksBEFG.
Nucleic Acids Res., 51, 2023
|
|
6VQL
 
 | |
6WYC
 
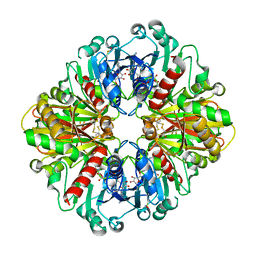 | |
6E21
 
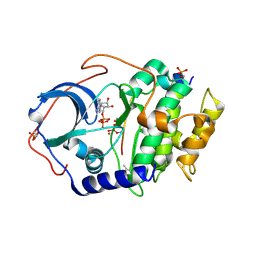 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-06 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
3RRD
 
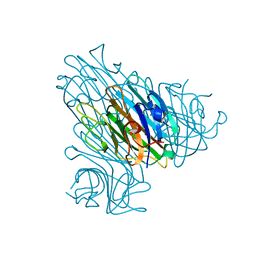 | | Native structure of Dioclea virgata lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION | | Authors: | Delatorre, P, Nobrega, R.B, Gadelha, C.A.A, Santi-Gadelha, T, Farias, D.L, Rocha, B.A.M, Cavada, B.S, Nagano, C.S, Bezerra, E.H.S, Bezerra, M.J, Alencar, K.L. | | Deposit date: | 2011-04-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production.
Biochimie, 94, 2012
|
|
6WKP
 
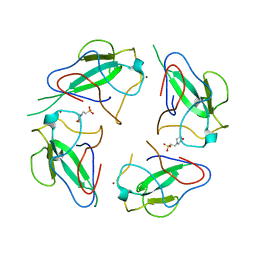 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
3RA0
 
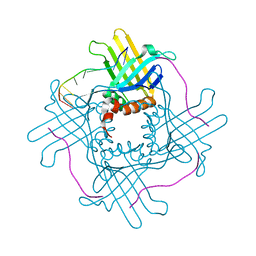 | |
3Q8G
 
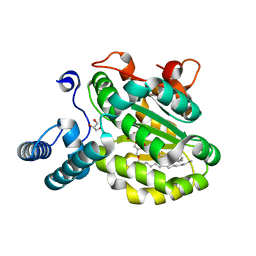 | | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CRAL-TRIO domain-containing protein YKL091C, GLYCEROL | | Authors: | Ortlund, E.A, Schaaf, G, Bankaitis, V.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution.
Mol.Biol.Cell, 22, 2011
|
|
7F3Y
 
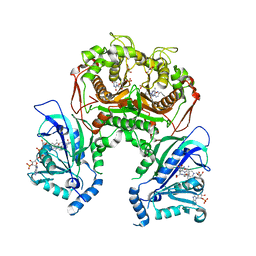 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
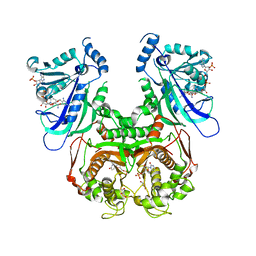 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
3R7Q
 
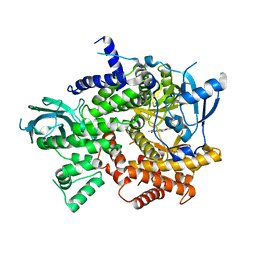 | | Structure-based design of thienobenzoxepin inhibitors of PI3- kinase | | Descriptor: | N-(2-chlorophenyl)-N-methyl-4H-thieno[3,2-c]chromene-2-carboxamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design of thienobenzoxepin inhibitors of PI3-kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R9Y
 
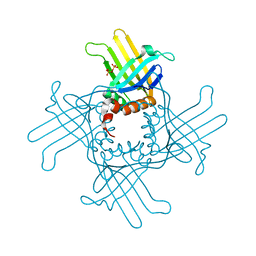 | | Crystal Structure of StWhy2 K67A (form I) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
3R7R
 
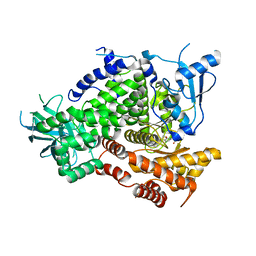 | | Structure-based design of thienobenzoxepin inhibitors of PI3-Kinase | | Descriptor: | 8-(acetylamino)-N-(2-chlorophenyl)-N-methyl-4,5-dihydrothieno[3,2-d][1]benzoxepine-2-carboxamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of thienobenzoxepin inhibitors of PI3-kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3O2M
 
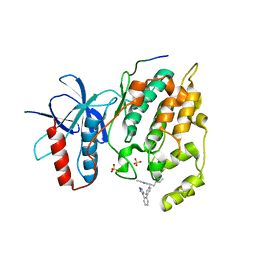 | |
3R9Z
 
 | | Crystal Structure of StWhy2 K67A (form II) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
