1C1H
 
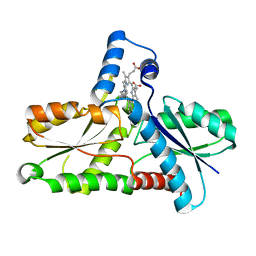 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS FERROCHELATASE IN COMPLEX WITH N-METHYL MESOPORPHYRIN | | Descriptor: | FERROCHELATASE, MAGNESIUM ION, N-METHYLMESOPORPHYRIN | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1D7F
 
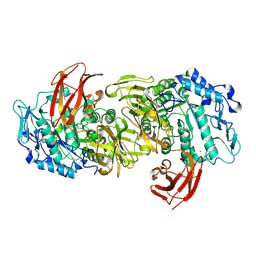 | | CRYSTAL STRUCTURE OF ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP. 1011 DETERMINED AT 1.9 A RESOLUTION | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-10-18 | | Release date: | 2000-03-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of asparagine 233-replaced cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 determined at 1.9 A resolution.
J.Mol.Recog., 13, 2000
|
|
1D7Q
 
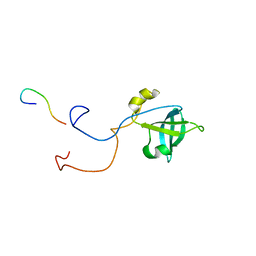 | | HUMAN TRANSLATION INITIATION FACTOR EIF1A | | Descriptor: | PROTEIN (N-TERMINAL HISTIDINE TAG), TRANSLATION INITIATION FACTOR 1A | | Authors: | Battiste, J.L, Pestova, T.V, Hellen, C.U.T, Wagner, G. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The eIF1A solution structure reveals a large RNA-binding surface important for scanning function.
Mol.Cell, 5, 2000
|
|
1GWX
 
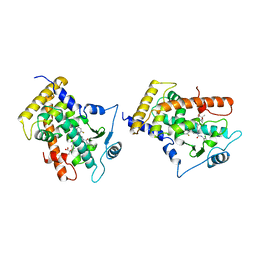 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3PDZ
 
 | |
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
1ECX
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1EKH
 
 | | NMR STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
1EKI
 
 | | AVERAGE SOLUTION STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
1DQX
 
 | | CRYSTAL STRUCTURE OF OROTIDINE 5'-PHOSPHATE DECARBOXYLASE COMPLEXED TO 6-HYDROXYURIDINE 5'-PHOSPHATE (BMP) | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Milburn, M.V, Miller, B.G, Hassell, A.M, Wolfenden, R, Short, S.A. | | Deposit date: | 2000-01-05 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anatomy of a proficient enzyme: the structure of orotidine 5'-monophosphate decarboxylase in the presence and absence of a potential transition state analog.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DQW
 
 | | CRYSTAL STRUCTURE OF OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Milburn, M.V, Miller, B.G, Hassell, A.M, Wolfenden, R, Short, S.A. | | Deposit date: | 2000-01-05 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anatomy of a proficient enzyme: the structure of orotidine 5'-monophosphate decarboxylase in the presence and absence of a potential transition state analog.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EGW
 
 | | CRYSTAL STRUCTURE OF MEF2A CORE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, ... | | Authors: | Santelli, E, Richmond, T.J. | | Deposit date: | 2000-02-17 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of MEF2A core bound to DNA at 1.5 A resolution.
J.Mol.Biol., 297, 2000
|
|
1D3A
 
 | | CRYSTAL STRUCTURE OF THE WILD TYPE HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, HALOPHILIC MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1CZ8
 
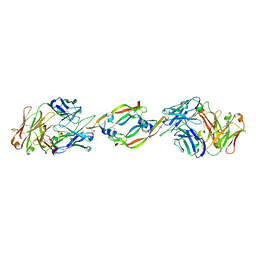 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH AN AFFINITY MATURED ANTIBODY | | Descriptor: | HEAVY CHAIN OF NEUTRALIZING ANTIBODY, LIGHT CHAIN OF NEUTRALIZING ANTIBODY, SULFATE ION, ... | | Authors: | Chen, Y, Wiesmann, C, Fuh, G, Li, B, Christinger, H.W, McKay, P, de Vos, A.M, Lowman, H.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-20 | | Last modified: | 2017-04-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured Fab in complex with antigen.
J.Mol.Biol., 293, 1999
|
|
1D8E
 
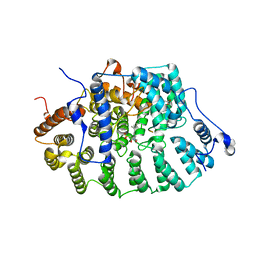 | | Zinc-depleted FTase complexed with K-RAS4B peptide substrate and FPP analog. | | Descriptor: | ACETATE ION, FARNESYLTRANSFERASE (ALPHA SUBUNIT), FARNESYLTRANSFERASE (BETA SUBUNIT), ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 1999-10-22 | | Release date: | 2000-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
Structure Fold.Des., 8, 2000
|
|
1DTQ
 
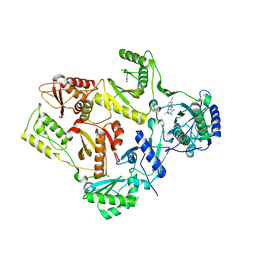 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1CE7
 
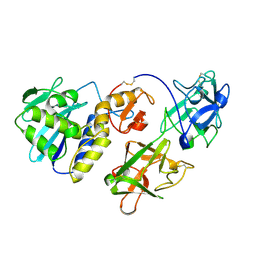 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
1CQR
 
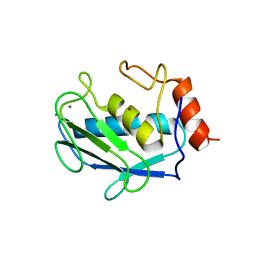 | | CRYSTAL STRUCTURE OF THE STROMELYSIN CATALYTIC DOMAIN AT 2.0 A RESOLUTION | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Chen, L, Rydel, T.J, Gu, F, Dunaway, C.M, Pikul, S, Dunham, K.M, Barnett, B.L. | | Deposit date: | 1999-08-11 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the stromelysin catalytic domain at 2.0 A resolution: inhibitor-induced conformational changes.
J.Mol.Biol., 293, 1999
|
|
1C9E
 
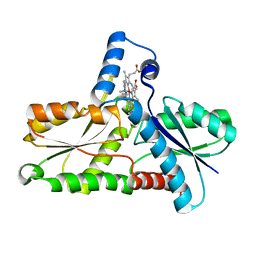 | | STRUCTURE OF FERROCHELATASE WITH COPPER(II) N-METHYLMESOPORPHYRIN COMPLEX BOUND AT THE ACTIVE SITE | | Descriptor: | MAGNESIUM ION, N-METHYLMESOPORPHYRIN CONTAINING COPPER, PROTOHEME FERROLYASE | | Authors: | Lecerof, D, Fodje, M.N, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1D2J
 
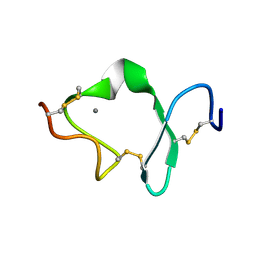 | | LDL RECEPTOR LIGAND-BINDING MODULE 6 | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | North, C.L, Blacklow, S.C. | | Deposit date: | 1999-09-23 | | Release date: | 2000-03-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sixth LDL-A module of the LDL receptor.
Biochemistry, 39, 2000
|
|
1C75
 
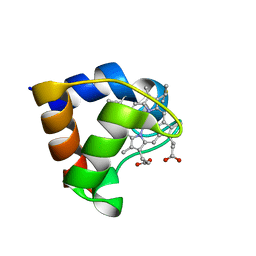 | | 0.97 A "AB INITIO" CRYSTAL STRUCTURE OF CYTOCHROME C-553 FROM BACILLUS PASTEURII | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
1C49
 
 | | STRUCTURAL AND FUNCTIONAL DIFFERENCES OF TWO TOXINS FROM THE SCORPION PANDINUS IMPERATOR | | Descriptor: | TOXIN K-BETA | | Authors: | Klenk, K.C, Tenenholz, T.C, Matteson, D.R, Rogowski, R.S, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1999-08-17 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural and functional differences of two toxins from the scorpion Pandinus imperator.
Proteins, 38, 2000
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DGJ
 
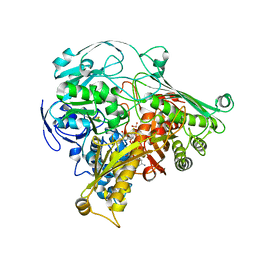 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
1CFZ
 
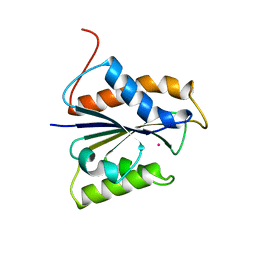 | | HYDROGENASE MATURATING ENDOPEPTIDASE HYBD FROM E. COLI | | Descriptor: | CADMIUM ION, HYDROGENASE 2 MATURATION PROTEASE | | Authors: | Fritsche, E, Paschos, A, Beisel, H.-G, Boeck, A, Huber, R. | | Deposit date: | 1999-03-23 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the hydrogenase maturating endopeptidase HYBD from Escherichia coli.
J.Mol.Biol., 288, 1999
|
|
