7JI1
 
 | |
7UPI
 
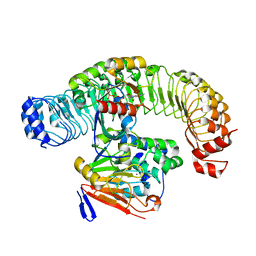 | | Cryo-EM structure of SHOC2-PP1c-MRAS holophosphatase complex | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, Leucine-rich repeat protein SHOC-2, ... | | Authors: | Fuller, J.R, Hajian, B, Lemke, C, Kwon, J, Bian, Y, Aguirre, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
6WO9
 
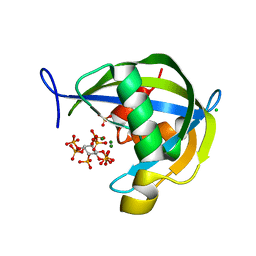 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1-diphosphoinositol pentakisphosphate (1-IP7) and Mg | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOG
 
 | |
6WOI
 
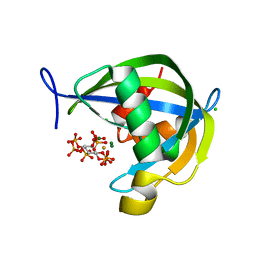 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1-Diphosphoinositol pentakisphosphate, Mg, and Fluoride ion, presoaked with 1,5-IP8, Mg and Fluoride for 30 seconds | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOE
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with inositol hexakisphosphate and Mg, presoaked with 5-IP7, Mg and Fluoride, soaking 10min in the absence of Fluoride. | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WO7
 
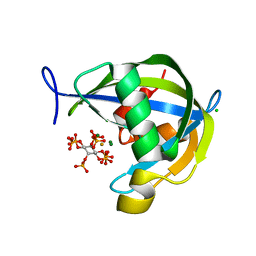 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-Diphosphoinositol pentakisphosphate (5-IP7), Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WO8
 
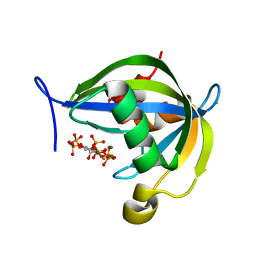 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5-PP-IP4), Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOA
 
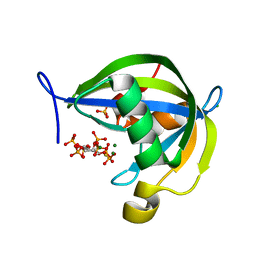 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 2-Diphosphoinositol pentakisphosphate (2-IP7), Mg, and Fluoride ion | | Descriptor: | (1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOD
 
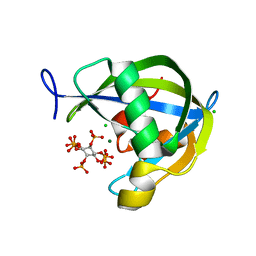 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with inositol hexakisphosphate and Mg, presoaked with 5-IP7, Mg and Fluoride, soaking 2min in the absence of Fluoride. | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
7UWT
 
 | | Structure of Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB_Vv F70A/F108Y (NTR 2.0) in complex with FMN at 1.85 Angstroms resolution | | Descriptor: | ACETATE ION, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sharrock, A.V, Arcus, V, Mumm, J.S, Ackerley, D.F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity.
Int J Mol Sci, 24, 2023
|
|
6WOF
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with inositol hexakisphosphate and Mg, presoaked with 5-IP7, Mg and Fluoride, soaking 20min in the absence of Fluoride. | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOB
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 4-diphosphoinositol pentakisphosphate (4-IP7), Mg, and Fluoride ion | | Descriptor: | (1S,2R,3S,4S,5S,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, B.S. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOC
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-diphosphoinositol pentakisphosphate and Mg, presoaked with 5-IP7, Mg and Fluoride, soaking 1min in the absence of Fluoride. | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6WOH
 
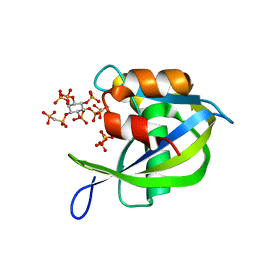 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1,5-di-methylenebisphosphonate inositol tetrakisphosphate (1,5-PCP-IP4) | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase 1, SULFATE ION, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
1H19
 
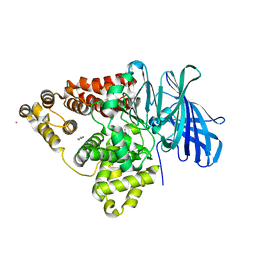 | | STRUCTURE OF [E271Q]LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2002-07-04 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Leukotriene A4 Hydrolase/Aminopeptidase, Glutamate 271 is a Catalyticresidue with Specific Roles in Two Distinct Enzyme Mechanisms
J.Biol.Chem., 277, 2002
|
|
1H7Z
 
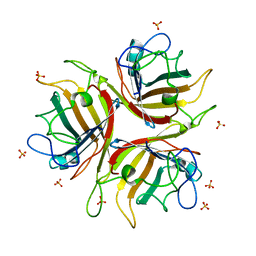 | | Adenovirus Ad3 fibre head | | Descriptor: | ADENOVIRUS FIBRE PROTEIN, SULFATE ION | | Authors: | Durmort, C, Stehlin, C, Schoehn, G, Mitraki, A, Drouet, E, Cusack, S, Burmeister, W.P. | | Deposit date: | 2001-01-21 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Fiber Head of Ad3, a Non-Car-Binding Serotype of Adenovirus
Virology, 285, 2001
|
|
1H5Y
 
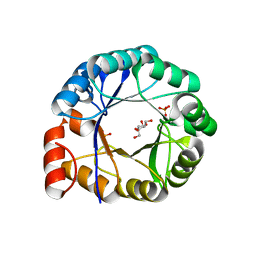 | | HisF protein from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, HISF, PHOSPHATE ION | | Authors: | Banfield, M.J, Lott, J.S, McCarthy, A.A, Baker, E.N. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7TQ8
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
1KZ5
 
 | |
8OIZ
 
 | | Crystal structure of human CRBN-DDB1 in complex with Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Le Bihan, Y.-V, Cabry, M.P, van Montfort, R.L.M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
7THU
 
 | | Structure of reduced bovine cytochrome c oxidase at 1.93 Angstrom resolution obtained by synchrotron X-rays | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
1HE1
 
 | | Crystal structure of the complex between the GAP domain of the Pseudomonas aeruginosa ExoS toxin and human Rac | | Descriptor: | ALUMINUM FLUORIDE, EXOENZYME S, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wurtele, M, Wolf, E, Pederson, K.J, Buchwald, G, Ahmadian, M.R, Barbieri, J.T, Wittinghofer, A. | | Deposit date: | 2000-11-18 | | Release date: | 2001-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Nat.Struct.Biol., 8, 2001
|
|
7TIH
 
 | | Structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
7TIE
 
 | | Structure of oxidized bovine cytochrome c oxidase at 1.90 Angstrom resolution obtained by synchrotron X-rays | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
