6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCR
 
 | | Crystal structure of E.coli RppH in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6CJ2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG056 | | Descriptor: | 2-{[3-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
Acs Chem.Biol., 13, 2018
|
|
6CXF
 
 | |
6TF7
 
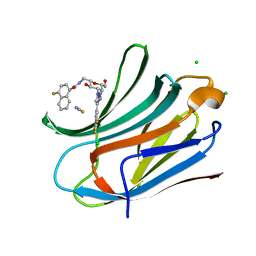 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
6F7U
 
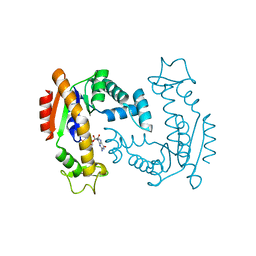 | | Molecular Mechanism of ATP versus GTP Selectivity of Adenylate Kinase | | Descriptor: | Adenylate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rogne, P, Rosselin, M, Grundstrom, C, Hedberg, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2017-12-12 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of ATP versus GTP selectivity of adenylate kinase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7FBD
 
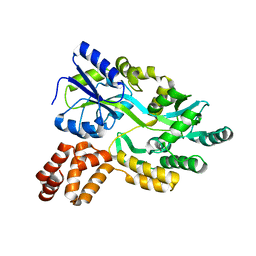 | | De novo design protein D53 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D53 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBB
 
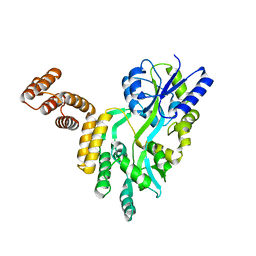 | | De novo design protein D12 with MBP tag | | Descriptor: | Maltodextrin-binding protein,de novo designed protein D12 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6HH5
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with ADP-HPM | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, GLYCEROL, ... | | Authors: | Ariza, A. | | Deposit date: | 2018-08-24 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
7FBC
 
 | | De novo design protein D22 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D22 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6HGZ
 
 | |
8STG
 
 | |
6CGD
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, Bifunctional AAC/APH, CHLORIDE ION, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
6CAV
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Dibekacin | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, Dibekacin, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
6HA4
 
 | | Crystal structure of PAF - p-sulfonatocalix[4]arene complex | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Pc24g00380 protein | | Authors: | Alex, J.M, Rennie, M, Engilberge, S, Batta, G, Crowley, P.B. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Calixarene-mediated assembly of a small antifungal protein.
Iucrj, 6, 2019
|
|
6T8C
 
 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in the apo form. | | Descriptor: | Formate dehydrogenase | | Authors: | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
6VPD
 
 | | Crystal structure of Trgpx in apo form | | Descriptor: | Glutathione peroxidase | | Authors: | Adriani, P.P, De Oliveira, G.S, Paiva, F.C.R, Dias, M.V.B, Chambergo, F.S. | | Deposit date: | 2020-02-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and functional characterization of the glutathione peroxidase-like thioredoxin peroxidase from the fungus Trichoderma reesei.
Int.J.Biol.Macromol., 167, 2020
|
|
8T9N
 
 | | Bacillus subtilis RsgI GGG mutant | | Descriptor: | Anti-sigma-I factor RsgI | | Authors: | Brogan, A.P, Habib, C, Hobbs, S.J, Kranzusch, P.J, Rudner, D.Z. | | Deposit date: | 2023-06-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial SEAL domains undergo autoproteolysis and function in regulated intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T6V
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6HKN
 
 | | Crystal structure of Compound 35 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [2-azanyl-4-(trifluoromethyloxy)phenyl]-[4-(7-methoxyquinazolin-4-yl)piperidin-1-yl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
6HOZ
 
 | |
8TAH
 
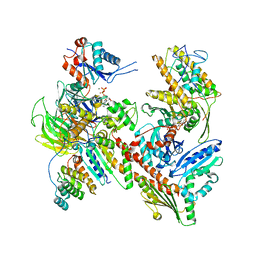 | | Cryo-EM structure of Cortactin-bound to Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | Fregoso, F.E, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of synergistic activation of Arp2/3 complex by cortactin and WASP-family proteins.
Nat Commun, 14, 2023
|
|
8T6U
 
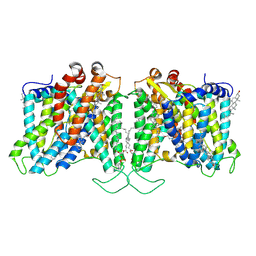 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7B8V
 
 | |
6TF6
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | CHLORIDE ION, Galectin-3, ~{N}-[[(2~{S},3~{S},4~{R},5~{S},6~{R})-4-[[5,6-bis(fluoranyl)-2-oxidanylidene-chromen-3-yl]methoxy]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]methyl]-4-fluoranyl-naphthalene-1-carboxamide | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
