8OQZ
 
 | | Structure of human gamma-secretase PSEN1 APH-1B isoform reconstituted into lipid nanodisc in complex with Ab46 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Odorcic, I, Chavez Gutierrez, L, Efremov, R.G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Apo and A beta 46-bound gamma-secretase structures provide insights into amyloid-beta processing by the APH-1B isoform.
Nat Commun, 15, 2024
|
|
7AEA
 
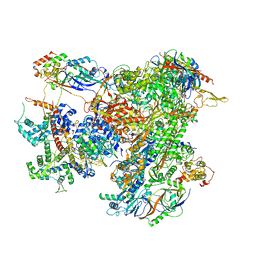 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1CMS
 
 | |
1JB5
 
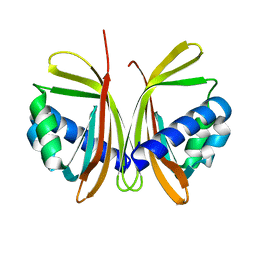 | | CRYSTAL STRUCTURE OF NTF2 M118E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1K19
 
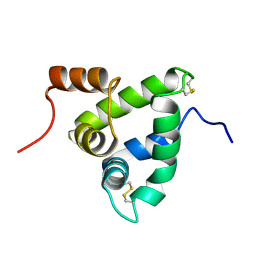 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
3P3A
 
 | |
1JFX
 
 | | Crystal structure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution | | Descriptor: | 1,4-beta-N-Acetylmuramidase M1, CHLORIDE ION | | Authors: | Rau, A, Hogg, T, Marquardt, R, Hilgenfeld, R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 A resolution.
J.Biol.Chem., 276, 2001
|
|
3OQ3
 
 | |
1DX9
 
 | | W57A Apoflavodoxin from Anabaena | | Descriptor: | Flavodoxin, SULFATE ION | | Authors: | Romero, A, Sancho, J. | | Deposit date: | 1999-12-23 | | Release date: | 2000-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissecting the Energetics of the Apoflavodoxin-Fmn Complex
J.Biol.Chem., 275, 2000
|
|
1JJU
 
 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JMX
 
 | |
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
1DQL
 
 | | CRYSTAL STRUCTURE OF AN UNLIGANDED (NATIVE) FV FROM A HUMAN IGM ANTI-PEPTIDE ANTIBODY | | Descriptor: | IGM MEZ IMMUNOGLOBULIN | | Authors: | Ramsland, P.A, Shan, L, Moomaw, C.R, Slaughter, C.A, Guddat, L.W, Edmundson, A.B. | | Deposit date: | 2000-01-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An unusual human IgM antibody with a protruding HCDR3 and high avidity for its peptide ligands.
Mol.Immunol., 37, 2000
|
|
1JB4
 
 | | CRYSTAL STRUCTURE OF NTF2 M102E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
8IW9
 
 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW4
 
 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
7AGJ
 
 | | Ribonucleotide Reductase R1 protein from Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
8IW7
 
 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ITF
 
 | | Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-22 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
1E1F
 
 | | Crystal structure of a Monocot (Maize ZMGlu1) beta-glucosidase in complex with p-Nitrophenyl-beta-D-thioglucoside | | Descriptor: | 4-nitrophenyl 1-thio-beta-D-glucopyranoside, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-05-03 | | Release date: | 2001-02-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Monocotyledon (Maize Zmglu1) Beta-Glucosidase and a Model of its Complex with P-Nitrophenyl Beta-D-Thioglucoside
Biochem.J., 354, 2001
|
|
7AL8
 
 | |
