3CSE
 
 | | Candida glabrata Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A.C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase.
Chem.Biol., 15, 2008
|
|
1JBE
 
 | |
2BS4
 
 | | GLU C180 -> ILE VARIANT QUINOL:FUMARATE REDUCTASE FROMWOLINELLA SUCCINOGENES | | Descriptor: | 2,3-DIMETHYL-1,4-NAPHTHOQUINONE, CITRIC ACID, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2005-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Experimental Support for the E-Pathway Hypothesis of Coupled Transmembrane Electron and Proton Transfer in Dihemic Quinol:Fumarate Reductase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5T5B
 
 | |
4R3W
 
 | | Crystal Structure Analysis of the 1,2,3-tricarboxylate benzoic acid bound to sp-ASADH-2'5'-ADP complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-2'-5'-DIPHOSPHATE, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2P9E
 
 | |
3TCY
 
 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum (cPAH) bound to phenylalanine in a site distal to the active site | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PHENYLALANINE, ... | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2011-08-09 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
1D4C
 
 | | CRYSTAL STRUCTURE OF THE UNCOMPLEXED FORM OF THE FLAVOCYTOCHROME C FUMARATE REDUCTASE OF SHEWANELLA PUTREFACIENS STRAIN MR-1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C FUMARATE REDUCTASE, HEME C, ... | | Authors: | Leys, D, Tsapin, A.S, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 1999-10-03 | | Release date: | 1999-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1.
Nat.Struct.Biol., 6, 1999
|
|
1IAY
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH COFACTOR PLP AND INHIBITOR AVG | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, 2-AMINO-4-(2-AMINO-ETHOXY)-BUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
2Z4X
 
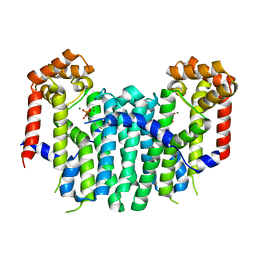 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-252 (P21) | | Descriptor: | (1-HYDROXYNONANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Chen, C.K.-M, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
4OE6
 
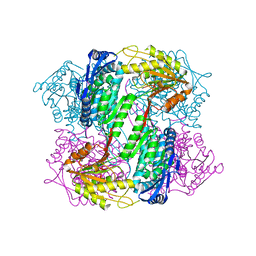 | | Crystal Structure of Yeast ALDH4A1 | | Descriptor: | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | | Authors: | Tanner, J.J. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Studies of Yeast Delta (1)-Pyrroline-5-carboxylate Dehydrogenase (ALDH4A1): Active Site Flexibility and Oligomeric State.
Biochemistry, 53, 2014
|
|
1QXS
 
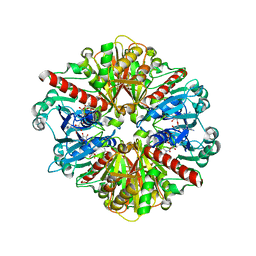 | | CRYSTAL STRUCTURE OF Trypanosoma cruzi GLYCERALDEHYDE-3- PHOSPHATE DEHYDROGENASE COMPLEXED WITH AN ANALOGUE OF 1,3- BisPHOSPHO-D-GLYCERIC ACID | | Descriptor: | 3-HYDROXY-2-OXO-4-PHOPHONOXY- BUTYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2003-09-08 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase complexed with an analogue of 1,3-bisphospho-d-glyceric acid.
Eur.J.Biochem., 270, 2003
|
|
2OUI
 
 | | D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Shimon, L, Burstein, Y, Goihberg, E, Peretz, M, Dym, O. | | Deposit date: | 2007-02-11 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
Proteins, 72, 2008
|
|
2DN1
 
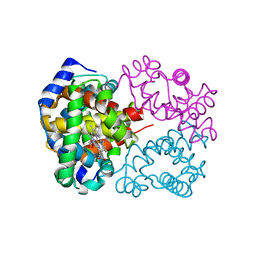 | | 1.25A resolution crystal structure of human hemoglobin in the oxy form | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, OXYGEN MOLECULE, ... | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
4JQK
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2-(2-aminopyridin-1-ium-1-yl)ethanol | | Descriptor: | 2-(2-aminopyridin-1-ium-1-yl)ethanol, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
5DYW
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)-N-(2-methoxyethyl)naphthalene-2-sulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coquelle, N, Brus, B, Colletier, J.P. | | Deposit date: | 2015-09-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
4G1R
 
 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Peculiarity in crystal packing of anti-HIV lectin actinohivin in complex with alpha (1-2)mannobiose.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K9V
 
 | | Complex of CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(3S,6S)-6-{[N-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-L-seryl]amino}octan-3-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
4D8F
 
 | | Chlamydia trachomatis NrdB with a Mn/Fe cofactor (procedure 1 - high Mn) | | Descriptor: | ACETIC ACID, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Dassama, L.M.K, Boal, A.K, Krebs, C, Rosenzweig, A.C, Bollinger Jr, J.M. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1.
J.Am.Chem.Soc., 134, 2012
|
|
1LLP
 
 | | LIGNIN PEROXIDASE (ISOZYME H2) PI 4.15 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Choinowski, T.H, Piontek, K, Glumoff, T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle.
J.Mol.Biol., 286, 1999
|
|
2IGY
 
 | | Achiral, Cheap and Potent Inhibitors of Plasmepsins II | | Descriptor: | N-[1-(3-METHYLBUTYL)PIPERIDIN-4-YL]-N-{4-[METHYL(PYRIDIN-4-YL)AMINO]BENZYL}-4-PENTYLBENZAMIDE, Plasmepsin-2 | | Authors: | Prade, L. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Achiral, Cheap, and Potent Inhibitors of Plasmepsins I, II, and IV.
Chemmedchem, 1, 2006
|
|
1LDN
 
 | | STRUCTURE OF A TERNARY COMPLEX OF AN ALLOSTERIC LACTATE DEHYDROGENASE FROM BACILLUS STEAROTHERMOPHILUS AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wigley, D.B, Gamblin, S.J, Turkenburg, J.P, Dodson, E.J, Piontek, K, Muirhead, H, Holbrook, J.J. | | Deposit date: | 1991-11-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution.
J.Mol.Biol., 223, 1992
|
|
4OE4
 
 | | Crystal Structure of Yeast ALDH4A1 Complexed with NAD+ | | Descriptor: | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanner, J.J. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural Studies of Yeast Delta (1)-Pyrroline-5-carboxylate Dehydrogenase (ALDH4A1): Active Site Flexibility and Oligomeric State.
Biochemistry, 53, 2014
|
|
3RMI
 
 | |
5DYT
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)-N-methylnaphthalene-2-sulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coquelle, N, Brus, B, Colletier, J.P. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
