3ZFJ
 
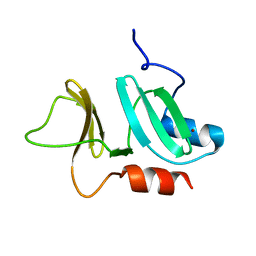 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
3ZFK
 
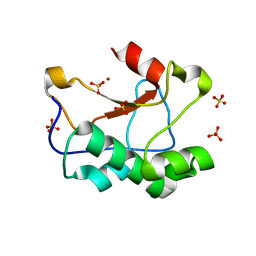 | | N-terminal truncated Nuclease Domain of Colicin E7 | | Descriptor: | ACETATE ION, CHLORIDE ION, COLICIN-E7, ... | | Authors: | Toth, E, Czene, A, Gyurcsik, B, Otten, H, Poulsen, J.-C.N, Larsen, S, Christensen, H.E.M, Nagata, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Insight Into the Zinc-Dependent DNA-Cleavage by the Colicin E7 Nuclease: A Crystallographic and Computational Study.
Metallomics, 6, 2014
|
|
3ZFL
 
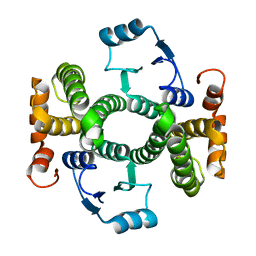 | | Crystal structure of the V58A mutant of human class alpha glutathione transferase in the apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Parbhoo, N, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Gildenhuys, S, Dirr, H.W. | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the V58A Mutant of Human Class Alpha Glutathione Transferase in the Apo Form
To be Published
|
|
3ZFM
 
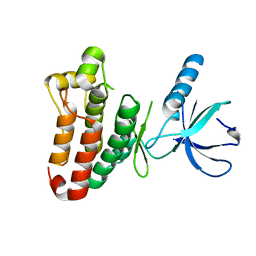 | | Crystal structure of EphB2 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZFN
 
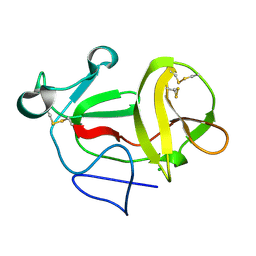 | | Crystal structure of product-like, processed N-terminal protease Npro | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFP
 
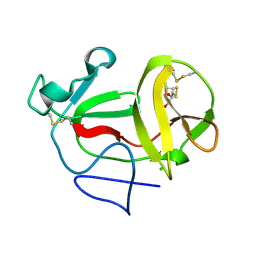 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFQ
 
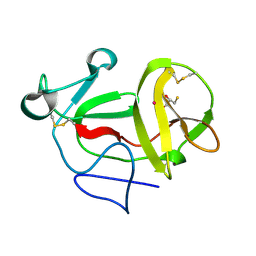 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | Descriptor: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFR
 
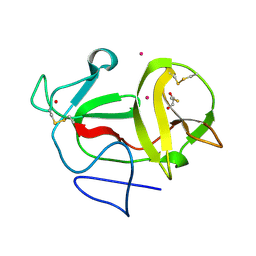 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | Descriptor: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFS
 
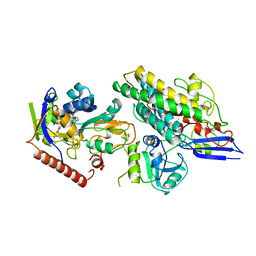 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
3ZFT
 
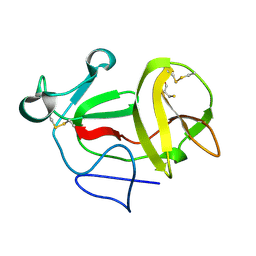 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFU
 
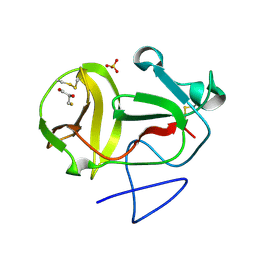 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | Descriptor: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFV
 
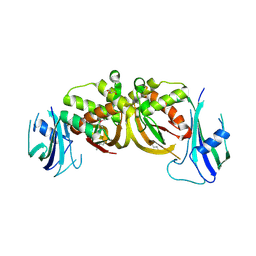 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
3ZFW
 
 | | Crystal structure of the TPR domain of kinesin light chain 2 in complex with a tryptophan-acidic cargo peptide | | Descriptor: | KINESIN LIGHT CHAIN 2, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY M MEMBER 2 | | Authors: | Pernigo, S, Lamprecht, A, Steiner, R.A, Dodding, M.P. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Kinesin-1:Cargo Recognition.
Science, 340, 2013
|
|
3ZFX
 
 | | Crystal structure of EphB1 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 1, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZFY
 
 | | Crystal structure of EphB3 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 3 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZFZ
 
 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by soaking | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZG0
 
 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by cocrystallization | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZG1
 
 | | NI-BOUND FORM OF M123A MUTANT OF CUPRIAVIDUS METALLIDURANS CH34 CNRXS | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Girard, E, Coves, J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Sensing and Signal Transduction by Cnrx from Cupriavidus Metallidurans Ch34: Role of the Only Methionine Assessed by a Functional, Spectroscopic, and Theoretical Study
Metallomics, 6, 2014
|
|
3ZG2
 
 | | Sterol 14 alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with the pyridine inhibitor (S)-2-(4-chlorophenyl)-2-(pyridin-3-yl)-1- (4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)ethanone (EPL-BS1246,UDO) | | Descriptor: | (S)-2-(4-chlorophenyl)-2-pyridin-3-yl-1-[4-[4-(trifluoromethyl)phenyl]piperazin-1-yl]ethanone, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
3ZG3
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH THE PYRIDINE INHIBITOR N-(1-(5-(trifluoromethyl)(pyridin-2-yl)) piperidin-4yl)-N-(4-(trifluoromethyl)phenyl)pyridin-3-amine (EPL- BS967, UDD) | | Descriptor: | N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)pyridin-2-yl]piperidin-4-yl]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
3ZG4
 
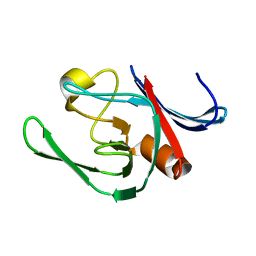 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZG5
 
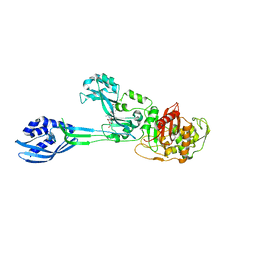 | | Crystal structure of PBP2a from MRSA in complex with peptidoglycan analogue at allosteric | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDOGLYCAN ANALOGUE, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZG6
 
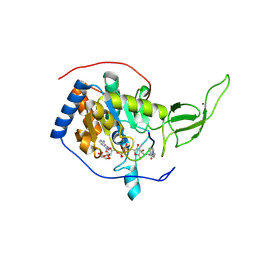 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
3ZG7
 
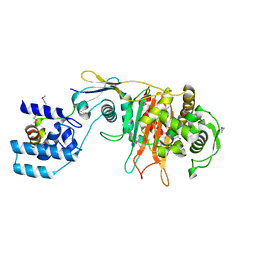 | |
