1R9K
 
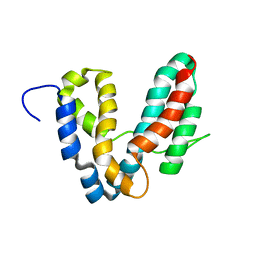 | | Representative solution structure of the catalytic domain of SopE2 | | Descriptor: | TypeIII-secreted protein effector: invasion-associated protein | | Authors: | Williams, C, Galyov, E.E, Bagby, S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Backbone Dynamics, and Interaction with Cdc42 of Salmonella Guanine Nucleotide Exchange Factor SopE2(,).
Biochemistry, 43, 2004
|
|
1RLQ
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RUW
 
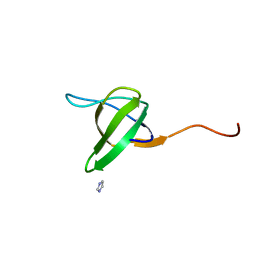 | | Crystal structure of the SH3 domain from S. cerevisiae Myo3 | | Descriptor: | IMIDAZOLE, Myosin-3 isoform | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the SH3 domain from S. cerevisiae Myo3
To be Published
|
|
1R6E
 
 | |
1RE0
 
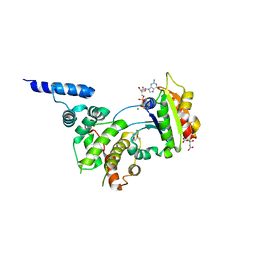 | | Structure of ARF1-GDP bound to Sec7 domain complexed with Brefeldin A | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-ribosylation factor 1, ARF guanine-nucleotide exchange factor 1, ... | | Authors: | Goldberg, J, Mossessova, E. | | Deposit date: | 2003-11-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism.
Mol.Cell, 12, 2003
|
|
4F7G
 
 | |
1S9D
 
 | | ARF1[DELTA 1-17]-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-Ribosylation Factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2004-02-04 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor
Nature, 426, 2003
|
|
4ETO
 
 | | Structure of S100A4 in complex with non-muscle myosin-IIA peptide | | Descriptor: | CALCIUM ION, Myosin-9, Protein S100-A4 | | Authors: | Ramagopal, U.A, Dulyaninova, N.G, Kumar, P.R, Almo, S.C, Bresnick, A.R, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of S100A4 with bound peptide P
To be Published
|
|
1TUU
 
 | | Acetate Kinase crystallized with ATPgS | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMMONIUM ION, ... | | Authors: | Gorrell, A, Lawrence, S.H, Ferry, J.G. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila.
J. Biol. Chem., 280, 2005
|
|
4HB4
 
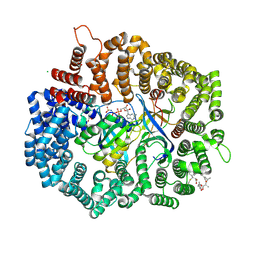 | |
4HAZ
 
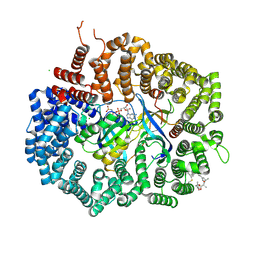 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(R543S,K548E,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1R8Q
 
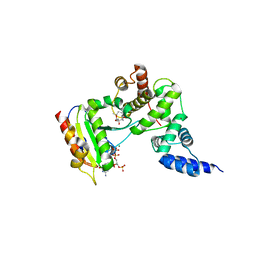 | | FULL-LENGTH ARF1-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-ribosylation factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor
Nature, 426, 2003
|
|
6SYT
 
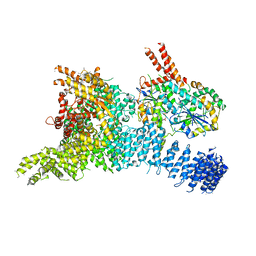 | | Structure of the SMG1-SMG8-SMG9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gat, Y, Schuller, J.M, Conti, E. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | InsP6binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4HAY
 
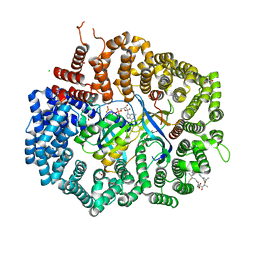 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K548E,K579Q)-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H0W
 
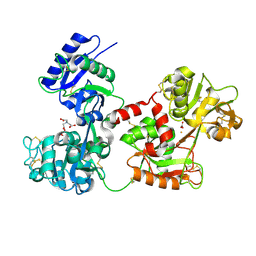 | | Bismuth bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bismuth(III) ION, CARBONATE ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2012-09-10 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe.
Sci Rep, 2, 2012
|
|
4HB3
 
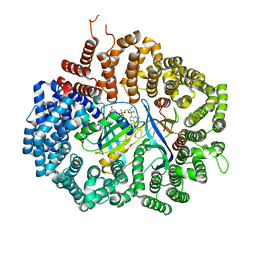 | |
4HAT
 
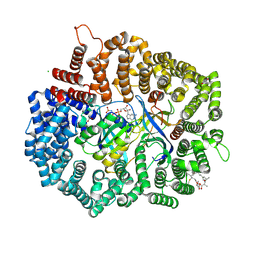 | |
4HAW
 
 | |
4EIK
 
 | |
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
4GPT
 
 | | Crystal structure of KPT251 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}ethyl)-1,3,4-oxadiazole, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells.
Leukemia, 27, 2013
|
|
1RLP
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RGP
 
 | | GTPASE-ACTIVATION DOMAIN FROM RHOGAP | | Descriptor: | RHOGAP | | Authors: | Barrett, T, Xiao, B, Dodson, E.J, Dodson, G, Ludbrook, S.B, Nurmahomed, K, Gamblin, S.J, Musacchio, A, Smerdon, S.J, Eccleston, J.F. | | Deposit date: | 1996-12-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the GTPase-activating domain from p50rhoGAP.
Nature, 385, 1997
|
|
1TUY
 
 | | Acetate Kinase complexed with ADP, AlF3 and acetate | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gorrell, A, Lawrence, S.H, Ferry, J.G. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Analyses of Arginine Residues in the Active Site of the Acetate Kinase from Methanosarcina thermophila.
J.Biol.Chem., 280, 2005
|
|
1U81
 
 | | Delta-17 Human ADP Ribosylation Factor 1 Complexed with GDP | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Seidel, R.D, Amor, J.C, Kahn, R.A, Prestegard, J.H. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational changes in human Arf1 on nucleotide exchange and deletion of membrane-binding elements.
J.Biol.Chem., 279, 2004
|
|
