4IJT
 
 | | Human p53 core domain with hot spot mutation R273H (form II) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
4IBQ
 
 | | Human p53 core domain with hot spot mutation R273C | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
2C9W
 
 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
2WNC
 
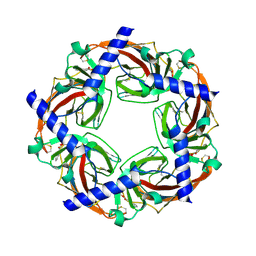 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
2YFO
 
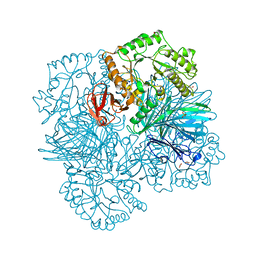 | | GALACTOSIDASE DOMAIN OF ALPHA-GALACTOSIDASE-SUCROSE KINASE, AGASK, in complex with galactose | | Descriptor: | ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
2WN9
 
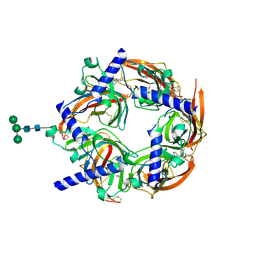 | | Crystal structure of Aplysia ACHBP in complex with 4-0H-DMXBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-5,6-DIHYDRO-2,3'-BIPYRIDIN-3(4H)-YLIDENEMETHYL]-3-METHOXYPHENOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
Embo J., 28, 2009
|
|
2WNL
 
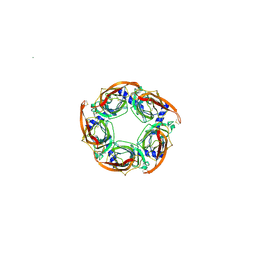 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH ANABASEINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4,5,6-tetrahydro-2,3'-bipyridine, 5-amino-1-pyridin-3-ylpentan-1-one, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
2WSP
 
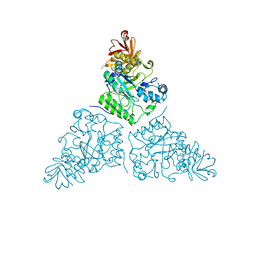 | | Thermotoga maritima alpha-L-fucosynthase, TmD224G, in complex with alpha-L-Fuc-(1-2)-beta-L-Fuc-N3 | | Descriptor: | ALPHA-L-FUCOSIDASE, PUTATIVE, alpha-L-fucopyranose-(1-2)-beta-L-fucosyl-azide | | Authors: | Sulzenbacher, G, Lipski, A, Cobucci-Ponzano, B, Conte, F, Bedini, E, Corsaro, M.M, Parrilli, M, Dal Piaz, F, Lepore, L, Rossi, M, Moracci, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Beta-Glycosyl Azides as Substrates for Alpha-Glycosynthases: Preparation of Novel Efficient Alpha-L-Fucosynthases
Chem.Biol., 16, 2009
|
|
5U5S
 
 | |
2YFN
 
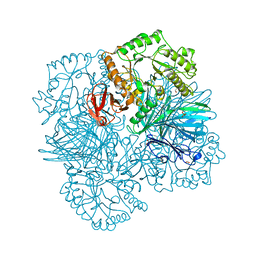 | | galactosidase domain of alpha-galactosidase-sucrose kinase, AgaSK | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
2WNJ
 
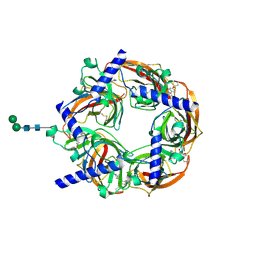 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH DMXBA | | Descriptor: | (3E)-3-[(2,4-DIMETHOXYPHENYL)METHYLIDENE]-3,4,5,6-TETRAHYDRO-2,3'-BIPYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants for Interaction of Partial Agonists with Acetylcholine Binding Protein and Neuronal Alpha7 Nicotinic Acetylcholine Receptor.
Embo J., 28, 2009
|
|
7WH8
 
 | |
7WHB
 
 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
7X2A
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
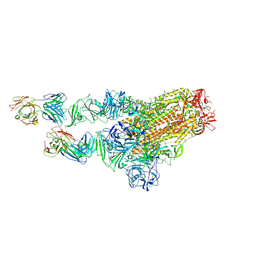 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X28
 
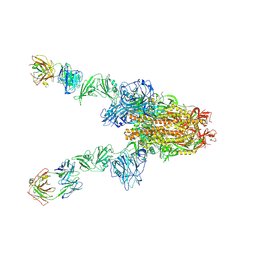 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7XTG
 
 | | Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1 | | Descriptor: | Bacteriocin curvacin-A, Mannose permease IIC component, Mannose permease IID component, ... | | Authors: | Zeng, J.W, Wang, J.W, Zhu, L.Y. | | Deposit date: | 2022-05-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural Basis of the Immunity Mechanisms of Pediocin-like Bacteriocins.
Appl.Environ.Microbiol., 88, 2022
|
|
7WHD
 
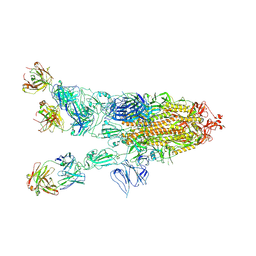 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
7WN6
 
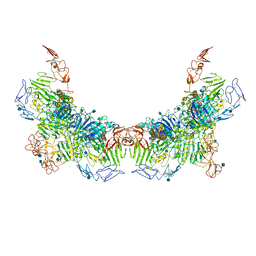 | | Cryo-EM structure of VWF D'D3 dimer (R1136M/E1143M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN4
 
 | | Cryo-EM structure of VWF D'D3 dimer (wild type) complexed with D1D2 at 3.4 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN3
 
 | | Cryo-EM structure of VWF D'D3 dimer (2M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
6KD0
 
 | | Crystal Structure of Vibralactone Cyclase | | Descriptor: | Vibralactone Cyclase | | Authors: | Zeng, Y, Feng, K.N. | | Deposit date: | 2019-06-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Hydrolase-Catalyzed Cyclization Forms the Fused Bicyclic beta-Lactone in Vibralactone.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6MNL
 
 | |
