4RZS
 
 | | Lac repressor engineered to bind sucralose, unliganded tetramer | | Descriptor: | GLYCEROL, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
6GCO
 
 | | Truncated FtsH from A. aeolicus in P312 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Uthoff, M, Baumann, U. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Conformational flexibility of pore loop-1 gives insights into substrate translocation by the AAA+protease FtsH.
J. Struct. Biol., 204, 2018
|
|
6GHL
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MALONATE ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5N2K
 
 | | Structure of unbound Briakinumab FAb | | Descriptor: | 1,2-ETHANEDIOL, Briakinumab FAb heavy chain, Briakinumab FAb light chain | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-02-07 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.219 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
8TRV
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be Published
|
|
8TV1
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_L1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
2K14
 
 | | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis | | Descriptor: | YuaF protein | | Authors: | Walker, C.A, Hinderhofer, M, Witte, D.J, Boos, W, Moller, H.M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis.
J.Biomol.Nmr, 42, 2008
|
|
6PEO
 
 | | Cryo-EM structure of alpha-synuclein H50Q Narrow Fibril | | Descriptor: | Alpha-synuclein | | Authors: | Boyer, D.R, Li, B, Sawaya, M.R, Jiang, L, Eisenberg, D.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8HZC
 
 | |
8HZB
 
 | |
8HZS
 
 | |
5FS7
 
 | |
4UHN
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | ACETIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UHM
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UHO
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
1I34
 
 | | SOLUTION DNA QUADRUPLEX WITH DOUBLE CHAIN REVERSAL LOOP AND TWO DIAGONAL LOOPS CONNECTING GGGG TETRADS FLANKED BY G-(T-T) TRIAD AND T-T-T TRIPLE | | Descriptor: | 5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*CP*AP*GP*GP*GP*TP*TP*TP*TP*GP*GP*T)-3' | | Authors: | Kuryavyi, V, Majumdar, A, Shallop, A, Chernichenko, N, Skripkin, E, Jones, R, Patel, D.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple.
J.Mol.Biol., 310, 2001
|
|
6HKA
 
 | |
6GHR
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GFQ
 
 | | cyanobacterial GAPDH with NAD and CP12 bound | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVE
 
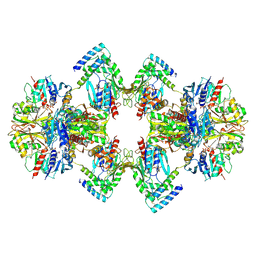 | | GAPDH-CP12-PRK complex | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Shah, N, Bubeck, D, Murray, J.W. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4M9K
 
 | |
4M9M
 
 | |
2HI9
 
 | |
