5X68
 
 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
3F4J
 
 | | Crystal structure of LeuT bound to glycine and sodium | | Descriptor: | GLYCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3WHA
 
 | | Hsp90 alpha N-terminal domain in complex with a tricyclic inhibitor | | Descriptor: | 4-{[4-amino-6-(5-chloro-1H,3H-benzo[de]isochromen-6-yl)-1,3,5-triazin-2-yl]sulfanyl}butanamide, GLYCEROL, Heat shock protein HSP 90-alpha, ... | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2013-08-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and synthesis of 2-amino-6-(1H,3H-benzo[de]isochromen-6-yl)-1,3,5-triazines as novel Hsp90 inhibitors
Bioorg.Med.Chem., 22, 2014
|
|
6YU6
 
 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU5
 
 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
2BQ2
 
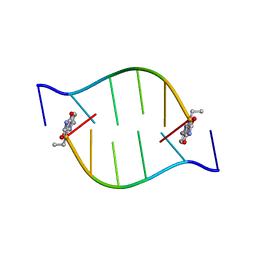 | | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*2AU)-3', NALIDIXIC ACID | | Authors: | Siegmund, K, Maheshwary, S, Narayanan, S, Connors, W, Richert, M. | | Deposit date: | 2005-04-26 | | Release date: | 2006-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it.
Nucleic Acids Res., 33, 2005
|
|
6C96
 
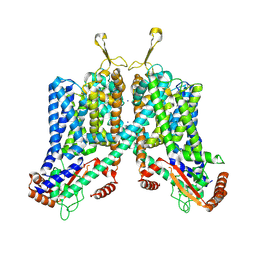 | | Cryo-EM structure of mouse TPC1 channel in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
6YU4
 
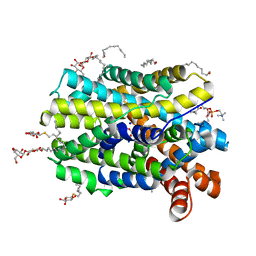 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU7
 
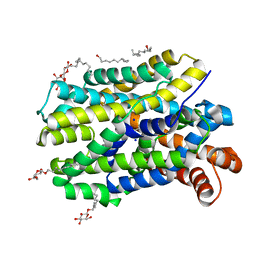 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
2QEI
 
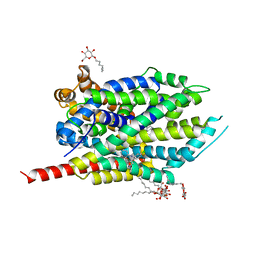 | | Crystal structure analysis of LeuT complexed with L-alanine, sodium, and clomipramine | | Descriptor: | 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ALANINE, SODIUM ION, ... | | Authors: | Singh, S.K, Yamashita, A, Gouaux, E. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antidepressant binding site in a bacterial homologue of neurotransmitter transporters.
Nature, 448, 2007
|
|
2QB4
 
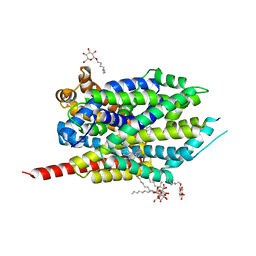 | | Crystal Structure Analysis of LeuT complexed with L-leucine, sodium and desipramine | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, LEUCINE, SODIUM ION, ... | | Authors: | Singh, S.K, Yamashita, A, Gouaux, E. | | Deposit date: | 2007-06-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antidepressant binding site in a bacterial homologue of neurotransmitter transporters.
Nature, 448, 2007
|
|
3BV9
 
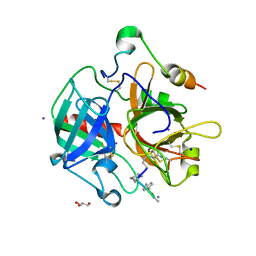 | | Structure of Thrombin Bound to the Inhibitor FM19 | | Descriptor: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | Authors: | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | Deposit date: | 2008-01-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
1GEN
 
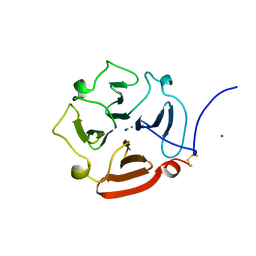 | | C-TERMINAL DOMAIN OF GELATINASE A | | Descriptor: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | Authors: | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
4NTY
 
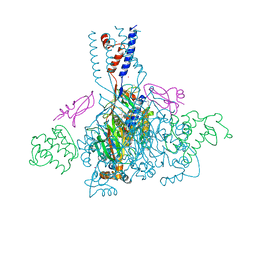 | | Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
1NHJ
 
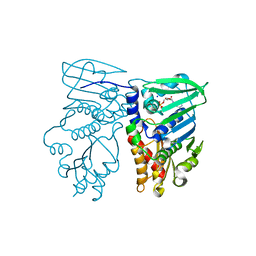 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | Descriptor: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
4P5M
 
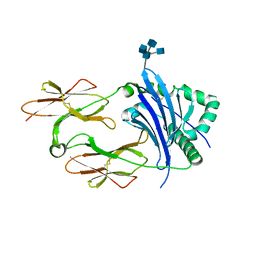 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergic Hypersensitivity and Autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S, Kappler, J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
1GVF
 
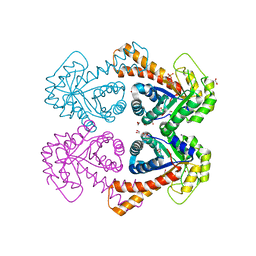 | | Structure of tagatose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOGLYCOLOHYDROXAMIC ACID, SODIUM ION, ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 2002-02-11 | | Release date: | 2002-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Tagatose-1,6-Bisphosphate Aldolase; Insight Into Chiral Discrimination, Mechanism and Specificity of Class II Aldolases
J.Biol.Chem., 277, 2002
|
|
2J5W
 
 | | Ceruloplasmin revisited: structural and functional roles of various metal cation binding sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CERULOPLASMIN, ... | | Authors: | Bento, I, Peixoto, C, Zaitsev, V.N, Lindley, P.F. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ceruloplasmin Revisited: Structural and Functional Roles of Various Metal Cation-Binding Sites.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1Y7W
 
 | | Crystal structure of a halotolerant carbonic anhydrase from Dunaliella salina | | Descriptor: | ACETIC ACID, Halotolerant alpha-type carbonic anhydrase (dCA II), SODIUM ION, ... | | Authors: | Premkumar, L, Greenblatt, H.M, Bageshwar, U.K, Savchenko, T, Gokhman, I, Sussman, J.L, Zamir, A, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-12-10 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Three-dimensional structure of a halotolerant algal carbonic anhydrase predicts halotolerance of a mammalian homolog.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2D10
 
 | |
1T3M
 
 | | Structure of the isoaspartyl peptidase with L-asparaginase activity from E. coli | | Descriptor: | NITRATE ION, Putative L-asparaginase, SODIUM ION | | Authors: | Prahl, A, Pazgier, M, Hejazi, M, Lockau, W, Lubkowski, J. | | Deposit date: | 2004-04-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2BEC
 
 | | Crystal structure of CHP2 in complex with its binding region in NHE1 and insights into the mechanism of pH regulation | | Descriptor: | Calcineurin B homologous protein 2, Sodium/hydrogen exchanger 1, YTTRIUM (III) ION | | Authors: | Ben Ammar, Y, Takeda, S, Hisamitsu, T, Mori, H, Wakabayashi, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation
Embo J., 25, 2006
|
|
