7A3D
 
 | |
7A2J
 
 | |
7A2V
 
 | |
7A37
 
 | |
7A2Q
 
 | |
7A33
 
 | |
7A2W
 
 | |
7A3A
 
 | |
7A2P
 
 | |
7A2N
 
 | |
7A2M
 
 | |
7A2R
 
 | |
7JTQ
 
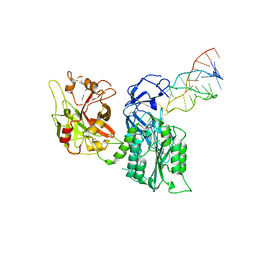 | |
7JTN
 
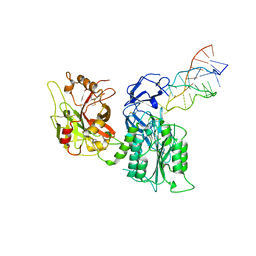 | |
7A2H
 
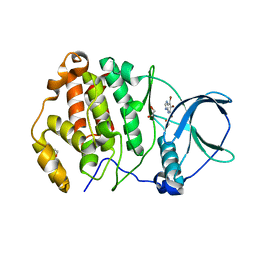 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-imidazo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7CTG
 
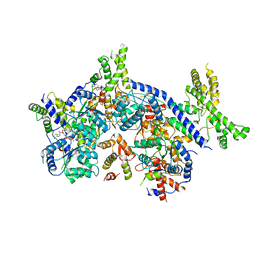 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
7JT9
 
 | |
7CSX
 
 | |
7CSR
 
 | | Structure of Ephexin4 R676L | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JSY
 
 | |
7JSX
 
 | | EPYC1(106-135) peptide-bound Rubisco | | Descriptor: | EPYC1, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, ... | | Authors: | Matthies, D, He, S, Jonikas, M.C. | | Deposit date: | 2020-08-16 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7CSL
 
 | | Crystal structure of the archaeal EF1A-EF1B complex | | Descriptor: | Elongation factor 1-alpha, Elongation factor 1-beta | | Authors: | Suzuki, T, Ito, K, Miyoshi, T, Murakami, R, Uchiumi, T. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the Switching Off of the Interaction between the Archaeal Ribosomal Stalk and aEF1A by Nucleotide Exchange Factor aEF1B.
J.Mol.Biol., 433, 2021
|
|
7A22
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7CSP
 
 | | Structure of Ephexin4 IDPSH | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
