3BSX
 
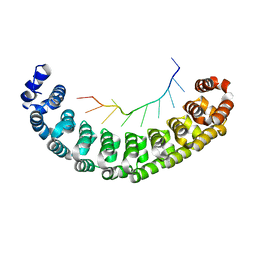 | | Crystal Structure of Human Pumilio 1 in complex with Puf5 RNA | | Descriptor: | 5'-R(*UP*UP*GP*UP*AP*AP*UP*AP*UP*UP*A)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
3HVO
 
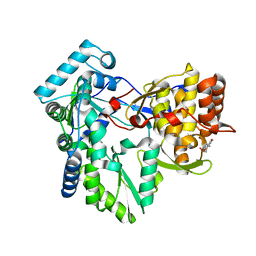 | | Structure of the genotype 2B HCV polymerase bound to a NNI | | Descriptor: | 2-(3-bromophenyl)-6-[(2-hydroxyethyl)amino]-1h-benzo[de]isoquinoline-1,3(2h)-dione, Genome polyprotein | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Biological Evaluation of a Series of 1H-Benzo[de]isoquinoline-1,3(2H)-diones as Hepatitis C Virus NS5B Polymerase Inhibitors
To be Published
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1JOX
 
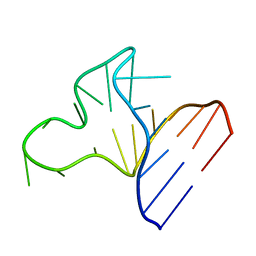 | |
3QJP
 
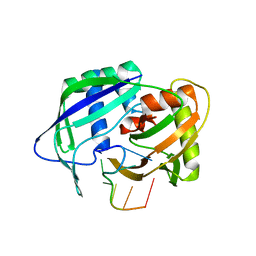 | |
6WHU
 
 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHW
 
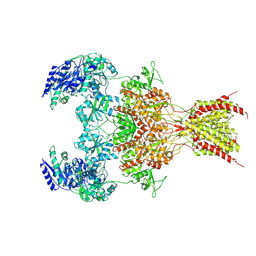 | | GluN1b-GluN2B NMDA receptor in complex with GluN2B antagonist SDZ 220-040, class 1 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHY
 
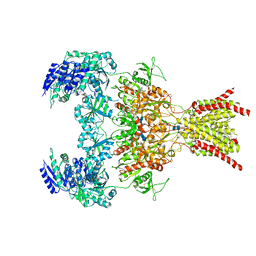 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
2RQA
 
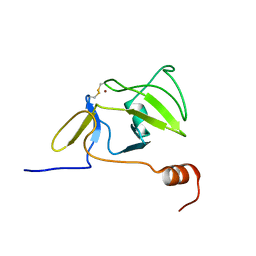 | | Solution structure of LGP2 CTD | | Descriptor: | ATP-dependent RNA helicase DHX58, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
2NBY
 
 | |
1AQO
 
 | | IRON RESPONSIVE ELEMENT RNA HAIRPIN, NMR, 15 STRUCTURES | | Descriptor: | IRON RESPONSIVE ELEMENT RNA HAIRPIN | | Authors: | Addess, K.J, Pardi, A. | | Deposit date: | 1997-07-31 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins.
J.Mol.Biol., 274, 1997
|
|
2MQP
 
 | |
2NBZ
 
 | |
2NC0
 
 | |
2NC1
 
 | |
4YE2
 
 | |
1CVJ
 
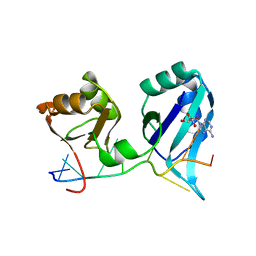 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
3QJL
 
 | |
3IVK
 
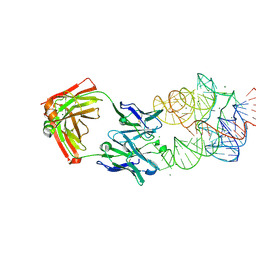 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
4NC7
 
 | | N-terminal domain of delta-subunit of RNA polymerase complexed with I3C and nickel ions | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
3QJJ
 
 | |
4NC8
 
 | | N-terminal domain of delta-subunit of RNA polymerase complexed with nickel ions | | Descriptor: | DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
1J26
 
 | | Solution structure of a putative peptidyl-tRNA hydrolase domain in a mouse hypothetical protein | | Descriptor: | immature colon carcinoma transcript 1 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality
J.Mol.Biol., 2010
|
|
1AUU
 
 | |
1JP0
 
 | |
