3IVP
 
 | |
443D
 
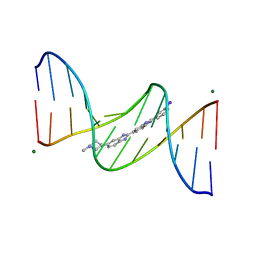 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'/ BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
2AC0
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1QUM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA | | Descriptor: | 5'-D(*(3DR)P*CP*GP*AP*CP*GP*A)-3', 5'-D(*CP*GP*TP*CP*C)-3', 5'-D(*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*G)-3', ... | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
2XQC
 
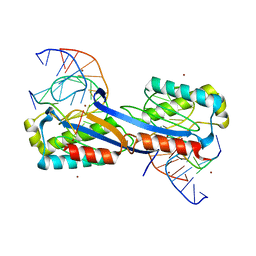 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
1DL8
 
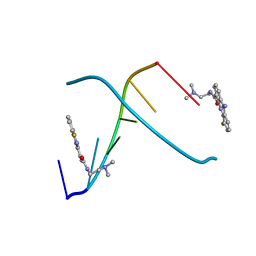 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
2ATA
 
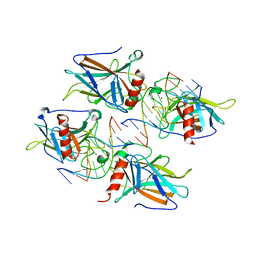 | | Structural Basis of DNA Recognition by p53 Tetramers (complex II) | | Descriptor: | 5'-D(*AP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*T)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-08-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1VJF
 
 | |
2MG8
 
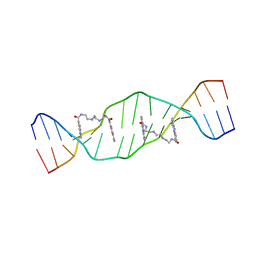 | |
3QK2
 
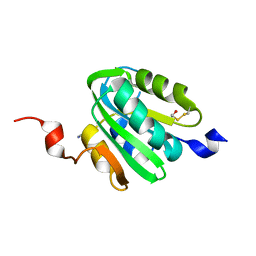 | |
3EBC
 
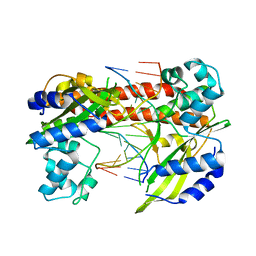 | | Structure of N141A HincII with Cognate DNA | | Descriptor: | 5'-D(*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DGP*DGP*DGP*DC)-3', MANGANESE (II) ION, ... | | Authors: | Little, E.J, Babic, A.C, Horton, N.C. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Early Interrogation and Recognition of DNA Sequence by Indirect Readout
Structure, 16, 2008
|
|
3HGD
 
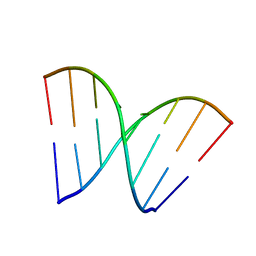 | |
4B3O
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LH6
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2VBL
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*DA*DA*DA*DA*DG*DG*DC*DA*DG*DAP)-3', 5'-D(*DA*DG*DG*DA*DT*DC*DC*DT*DA*DAP)-3', 5'-D(*DT*DC*DT*DG*DC*DC*DT*DT*DT*DT*DT*DT *DGP*DAP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
1KC6
 
 | | HincII Bound to Cognate DNA | | Descriptor: | 5'-D(P*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*G)-3', SODIUM ION, TYPE II RESTRICTION ENZYME HINCII | | Authors: | Horton, N.C, Dorner, L.F, Perona, J.J. | | Deposit date: | 2001-11-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation.
Nat.Struct.Biol., 9, 2002
|
|
2VBN
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*AP*AP*AP*AP*GP*GP*CP*AP*GP*AP)-3', 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*AP*AP)-3', 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
4DOG
 
 | |
3BAA
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-{[2-(1H-imidazol-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
1KF1
 
 | |
4LH7
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-aminothieno[3,2-c]pyridine-2,7-dicarboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Boriack-Sjodin, P.A, Prince, D.B. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1ZGW
 
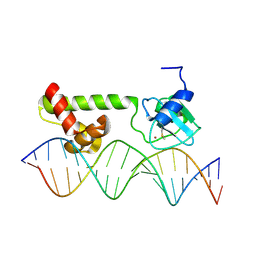 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
3BA8
 
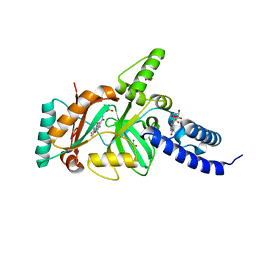 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 2-amino-7-fluoro-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAB
 
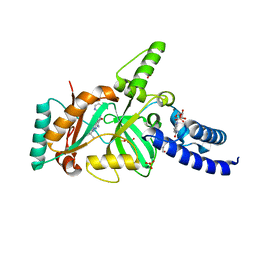 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-(4-pyrimidin-2-ylpiperazin-1-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3SC3
 
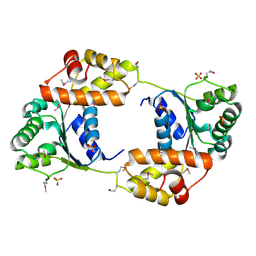 | |
