5WL2
 
 | | VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Descriptor: | Germline-reverted light chain of CR9114, Heavy chain of VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem.
To Be Published
|
|
6QQ8
 
 | |
5WLL
 
 | |
4UT4
 
 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | Descriptor: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
6QPM
 
 | | Adenovirus serotype 10 Fiber-Knob | | Descriptor: | Fiber protein | | Authors: | Baker, A.T, Rizkallah, P.J. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
6QRZ
 
 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
6QPT
 
 | | Crystal structure of nitrite bound synthetic core domain of nitrite reductase from Ralstonia pickettii (residues 1-331) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Antonyuk, S.V, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
8SKU
 
 | | Structure of human SIgA1 in complex with human CD89 (FcaR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Liu, Q, Stadtmueller, B.M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SIgA structures bound to Streptococcus pyogenes M4 and human CD89 provide insights into host-pathogen interactions.
Nat Commun, 14, 2023
|
|
6QSI
 
 | |
5KKV
 
 | |
1EHL
 
 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
6QRU
 
 | | X-ray radiation dose series on xylose isomerase - 2.01 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QTM
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | Descriptor: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | Authors: | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-18 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
4V7C
 
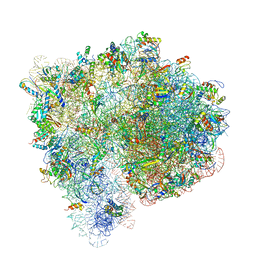 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6QTS
 
 | |
6DOH
 
 | |
6QSN
 
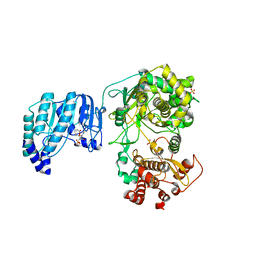 | | Crystal structure of Yellow fever virus polymerase NS5A | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Boura, E, Dubankova, A. | | Deposit date: | 2019-02-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses.
Antiviral Res., 169, 2019
|
|
6QU0
 
 | | Structure of azoreductase from Bacillus sp. A01 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, GLYCEROL, ... | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Crystallographic Studies of Azoreductase AzoA fromBacillus wakoensisA01.
Acs Chem.Biol., 15, 2020
|
|
6QU7
 
 | | Crystal structure of human DHODH in complex with BAY 2402234 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Friberg, A, Gradl, S. | | Deposit date: | 2019-02-26 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies.
Leukemia, 33, 2019
|
|
4W1V
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a thiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
6QUC
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUU
 
 | | Crystal Structure of KRAS-G12D in complex with GMP-PCP | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Fischer, G, Kessler, D, Muellauer, B, Wolkerstorfer, B. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | KRAS Binders Hidden in Nature.
Chemistry, 25, 2019
|
|
6QUT
 
 | |
5KTG
 
 | |
