6D2Z
 
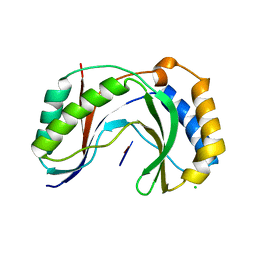 | | Structure of human Usb1 with uridine-adenosine, inactive H208Q mutant | | Descriptor: | 5'-R(UP*A)-3'), CHLORIDE ION, U6 snRNA phosphodiesterase | | Authors: | Nomura, Y, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-04-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural and mechanistic basis for preferential deadenylation of U6 snRNA by Usb1.
Nucleic Acids Res., 46, 2018
|
|
4N49
 
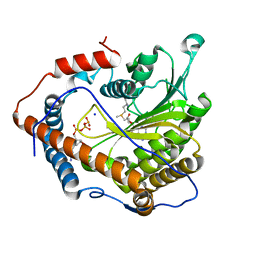 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
1HQM
 
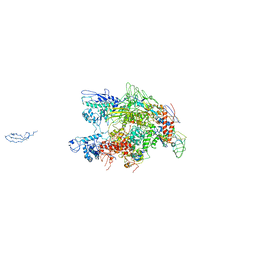 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2L94
 
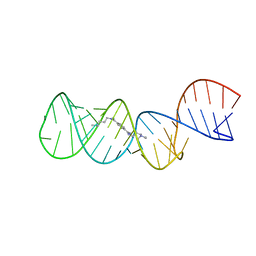 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
3V71
 
 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
8K20
 
 | | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | | Descriptor: | At4g34412, At5g53043, FE (III) ION, ... | | Authors: | Zheng, X.X, Zhu, L, Duan, L, Zhang, W.H. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Nucleic Acids Res., 52, 2024
|
|
3LY5
 
 | | DDX18 dead-domain | | Descriptor: | ATP-dependent RNA helicase DDX18, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-26 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2MDR
 
 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1SZY
 
 | |
1FOY
 
 | | THE RNA BINDING DOMAIN OF RIBOSOMAL PROTEIN L11: THREE-DIMENSIONAL STRUCTURE OF THE RNA-BOUND FORM OF THE PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RIBOSOMAL PROTEIN L11 | | Authors: | Hinck, A.P, Markus, M.A, Huang, S, Grzesiek, S, Kustanovich, I, Draper, D.E, Torchia, D.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA.
J.Mol.Biol., 274, 1997
|
|
1HWQ
 
 | |
3K4G
 
 | |
1Z3A
 
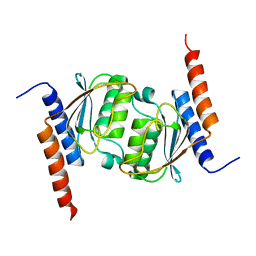 | | Crystal structure of tRNA adenosine deaminase TadA from Escherichia coli | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase | | Authors: | Malashkevich, V, Kim, J, Lisbin, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase.
Biochemistry, 45, 2006
|
|
4GWQ
 
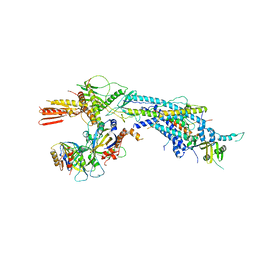 | | Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KL5
 
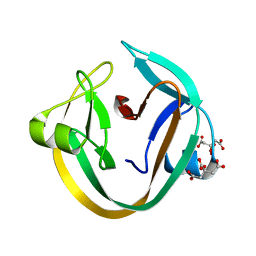 | | Crystal structure of NpuDnaE intein | | Descriptor: | CITRIC ACID, DNA polymerase III, alpha subunit, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
439D
 
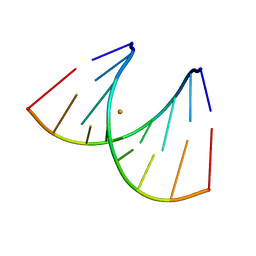 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4KL6
 
 | | Crystal structure of dimeric form of NpuDnaE intein | | Descriptor: | DNA-directed DNA polymerase,Nucleic acid binding, OB-fold, tRNA/helicase-type | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
2FD0
 
 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
2LMC
 
 | |
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6AIC
 
 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5U3G
 
 | | Structure of the Dickeya dadantii ykkC riboswitch bound to guanidinium | | Descriptor: | GUANIDINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Battaglia, R.A, Price, I.R, Ke, A. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for guanidine sensing by the ykkC family of riboswitches.
RNA, 23, 2017
|
|
1B42
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-1-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4GGK
 
 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) bound to tungstate | | Descriptor: | Mitochondrial cardiolipin hydrolase, TUNGSTATE(VI)ION, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
2HGH
 
 | |
