3ZKJ
 
 | | Crystal Structure of Ankyrin Repeat and Socs Box-Containing Protein 9 (Asb9) in Complex with Elonginb and Elonginc | | Descriptor: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Guo, K, Zhang, Y, Ayinampudi, V, Savitsky, P, Keates, T, Filippakopoulos, P, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, von Delft, F, Knapp, S, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2013-01-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
3AOP
 
 | | SULFITE REDUCTASE HEMOPROTEIN PHOTOREDUCED WITH PROFLAVINE EDTA, SIROHEME FEII,[4FE-4S] +1, PHOSPHATE BOUND | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-09 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
5VZP
 
 | | Sperm whale myoglobin H64Q with nitric oxide | | Descriptor: | GLYCEROL, Myoglobin, NITRIC OXIDE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
3FC1
 
 | | Crystal structure of p38 kinase bound to pyrimido-pyridazinone inhibitor | | Descriptor: | 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one, CHLORIDE ION, Mitogen-activated protein kinase 14 | | Authors: | Jacobs, M.D, Bellon, S. | | Deposit date: | 2008-11-20 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evaluating the molecular mechanics poisson-boltzmann surface area free energy method using a congeneric series of ligands to p38 MAP kinase.
J.Med.Chem., 48, 2005
|
|
3G81
 
 | |
3GOM
 
 | | Barium bound to the Holliday junction sequence d(TCGGCGCCGA)4 | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION | | Authors: | Naseer, A, Cardin, C.J. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1SKJ
 
 | | COCRYSTAL STRUCTURE OF UREA-SUBSTITUTED PHOSPHOPEPTIDE COMPLEX | | Descriptor: | 4-[3-CARBOXYMETHYL-3-(4-PHOSPHONOOXY-BENZYL)-UREIDO]-4-[(3-CYCLOHEXYL-PROPYL)-METHYL-CARBAMOYL]BUTYRIC ACID, PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and cocrystal structure of a nonpeptide Src SH2 domain ligand.
J.Med.Chem., 40, 1997
|
|
7OPQ
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CA1 in C188 | | Descriptor: | 1-[(2~{S})-2-(4-methoxyphenyl)pyrrolidin-1-yl]propan-1-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPP
 
 | | Crystal structure of the Rab27a fusion with Slp2a-RBDa1 effector for SF4 pocket drug targeting | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Synaptotagmin-like protein 2,Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
5LVH
 
 | |
6A05
 
 | | Structure of pSTING complex | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
5LVJ
 
 | |
5M24
 
 | | RARg mutant-S371E | | Descriptor: | (9cis)-retinoic acid, CHLORIDE ION, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Rochel, N, Sirigu, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Allosteric Regulation in the Ligand Binding Domain of Retinoic Acid Receptor gamma.
PLoS ONE, 12, 2017
|
|
1WRY
 
 | | Solution structure of the SH3 domain-binding glutamic acid-rich-like protein | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Inoue, K, Miyamoto, K, Nagashima, T, Hayashi, F, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-29 | | Release date: | 2005-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain-binding glutamic acid-rich-like protein
To be Published
|
|
7OVH
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 14 (JMV-6931) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVE
 
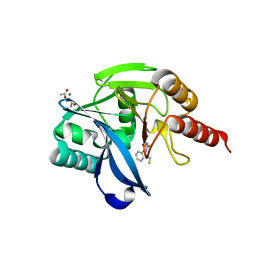 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 10 (JMV-7210) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Mangani, S, Docquier, J.D, Pozzi, C, Marcoccia, F. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OML
 
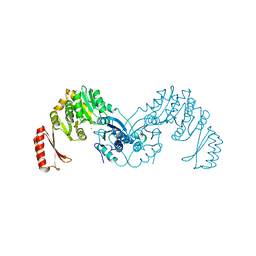 | |
7OLH
 
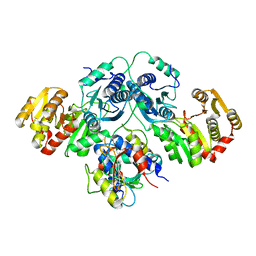 | |
7OJR
 
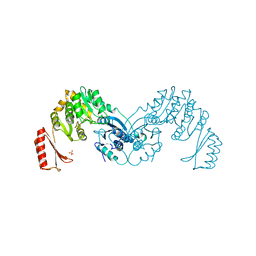 | |
7OJS
 
 | |
1A6M
 
 | | OXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
3D0F
 
 | | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Penicillin-binding 1 transmembrane protein MrcA | | Authors: | Cuff, M.E, Mulligan, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718.
TO BE PUBLISHED
|
|
8WRB
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
1B6Z
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-pyruvoyl tetrahydropterin synthase, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-18 | | Release date: | 2000-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
