5JQE
 
 | | Crystal structure of caspase8 tDED | | Descriptor: | Sugar ABC transporter substrate-binding protein,Caspase-8 chimera | | Authors: | Fu, T, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol. Cell, 64, 2016
|
|
4XAJ
 
 | | Crystal structure of human NR2E1/TLX | | Descriptor: | Atrophin/grunge, Maltose-binding periplasmic protein,Nuclear receptor subfamily 2 group E member 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
7VQO
 
 | | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | | Descriptor: | Ams1, Nbr1 and malE fusion protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1
Nat Commun, 13, 2022
|
|
4XR8
 
 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
5JON
 
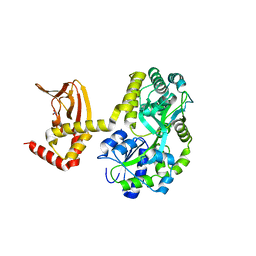 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
5JST
 
 | | MBP fused MDV1 coiled coil | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein,Mitochondrial division protein 1, ... | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | ACCORD: an assessment tool to determine the orientation of homodimeric coiled-coils.
Sci Rep, 7, 2017
|
|
4XA2
 
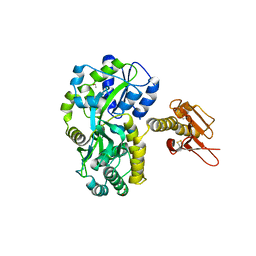 | |
5KBF
 
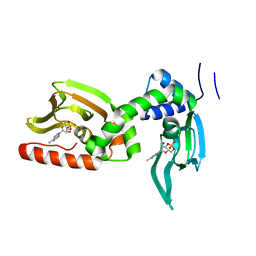 | | cAMP bound PfPKA-R (141-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit, putative | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
6PFO
 
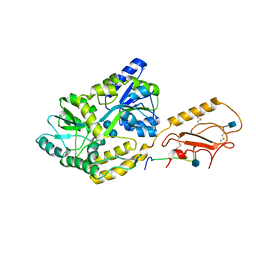 | |
5K94
 
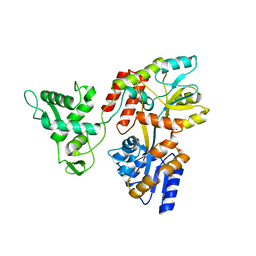 | | Deletion-Insertion Chimera of MBP with the Preprotein Cross-Linking Domain of the SecA ATPase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltose-binding periplasmic protein,Protein translocase subunit SecA,Maltose-binding periplasmic protein | | Authors: | Shilton, B.H, Hackett, J, Ghonaim, N. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a polypeptide-binding site in the DEAD Motor of the SecA ATPase.
FEBS Lett., 591, 2017
|
|
4XWO
 
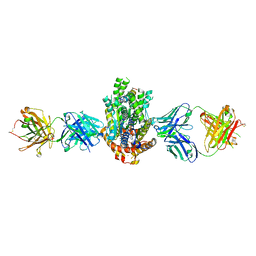 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
6PMI
 
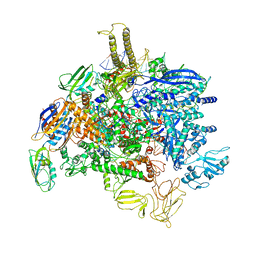 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
8IIZ
 
 | |
8IIY
 
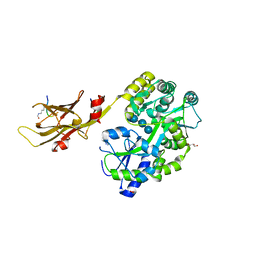 | |
8PZH
 
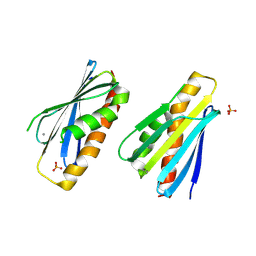 | | LpdD (H61A) mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of LpdD (H61A) mutant from Lactobacillus plantarum.
To Be Published
|
|
7WB2
 
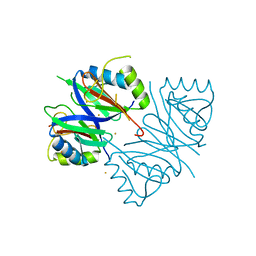 | | Oxidase ChaP-D49L/Y109F mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Zong, Y, Zheng, W, Wang, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8ILD
 
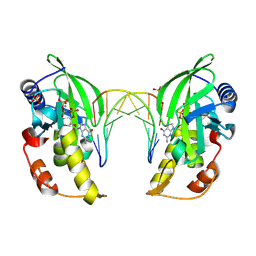 | | The crystal structure of native dGTP:DNApre-I:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*AP*TP*CP*CP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Qin, T, Chen, Y.Q, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILH
 
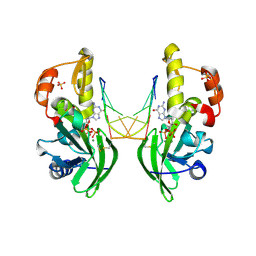 | | The crystal structure of dG(Se-Sp)-DNA:Pol X product binary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOSELENOPHOSPHATE, DNA (5'-D(*CP*GP*GP*AP*TP*CP*C)-3'), MAGNESIUM ION, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
8ILI
 
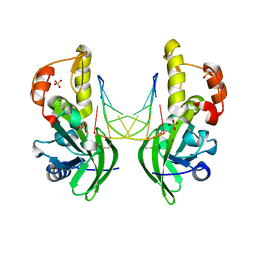 | | The crystal structure of dG(Se-Rp)-DNA:Pol X product binary complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*TP*CP*CP*(7S8))-3'), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
6QF7
 
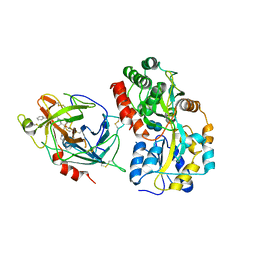 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1MH4
 
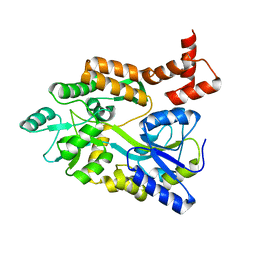 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
6Q62
 
 | |
4XZS
 
 | | Crystal Structure of TRIAP1-MBP fusion | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Abid-Ali, F, Perez-Dorado, I, Matthews, S.J. | | Deposit date: | 2015-02-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
1NL5
 
 | | Engineered High-affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
8RDV
 
 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2). | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
