5DTU
 
 | |
5ELX
 
 | | S. cerevisiae Dbp5 bound to RNA and mant-ADP BeF3 | | Descriptor: | ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Merchant, M.K, Modis, Y. | | Deposit date: | 2015-11-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5).
J.Mol.Biol., 428, 2016
|
|
5LTK
 
 | |
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
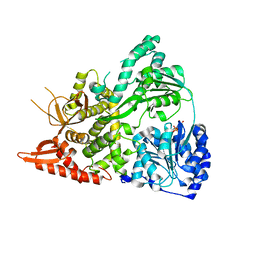
6UP4
 
 | |
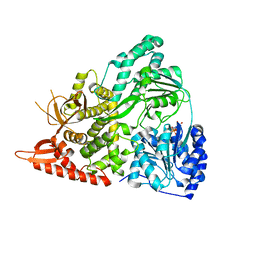
6UP3
 
 | |
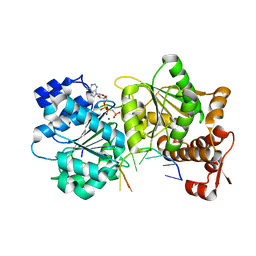
3SQW
 
 | |
3TBK
 
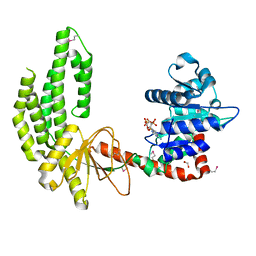 | | Mouse RIG-I ATPase Domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RIG-I Helicase Domain | | Authors: | Civril, F, Bennett, M.D, Hopfner, K.-P. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Embo Rep., 12, 2011
|
|
2EYQ
 
 | |
4F91
 
 | | Brr2 Helicase Region | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OOQ
 
 | | Structure of the Mtr4 Nop53 Complex | | Descriptor: | ATP-dependent RNA helicase DOB1, Ribosome biogenesis protein NOP53, SULFATE ION | | Authors: | Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53.
RNA, 23, 2017
|
|
4F92
 
 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F93
 
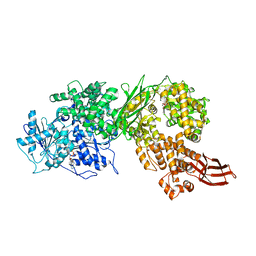 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5SUQ
 
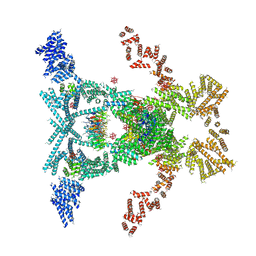 | | Crystal structure of the THO-Sub2 complex | | Descriptor: | 12-TUNGSTOPHOSPHATE, ATP-dependent RNA helicase SUB2, Tex1, ... | | Authors: | Ren, Y, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
5SUP
 
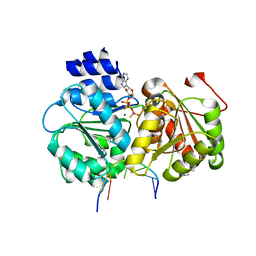 | | Crystal structure of the Sub2-Yra1 complex in association with RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase SUB2, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ren, Y, Schmiege, P, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
4XGT
 
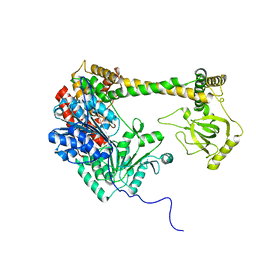 | |
6KYV
 
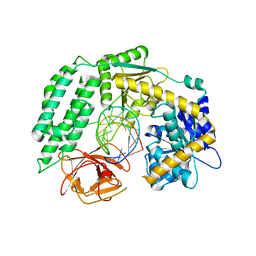 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
1S2M
 
 | |
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
1OYW
 
 | |
1OYY
 
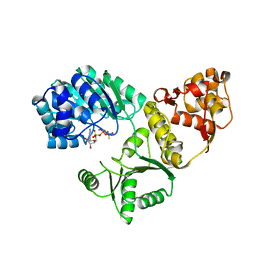 | | Structure of the RecQ Catalytic Core bound to ATP-gamma-S | | Descriptor: | ATP-dependent DNA helicase, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bernstein, D.A, Zittel, M.C, Keck, J.L. | | Deposit date: | 2003-04-07 | | Release date: | 2003-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution structure of the E. coli RecQ helicase catalytic core
Embo J., 22, 2003
|
|
1FUU
 
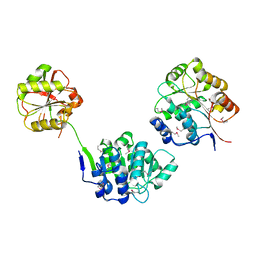 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1GKU
 
 | |
