5YDW
 
 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7DMW
 
 | | Crystal structure of CcpC regulatory domain in complex with citrate from Bacillus amyloliquefaciens | | Descriptor: | CITRATE ANION, CcpC | | Authors: | Chen, J, Wang, L, Shang, F, Liu, W, Chen, Y, Lan, J, Bu, T, Bai, X, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural analysis of catabolite control protein C that responds to citrate.
Sci Rep, 11, 2021
|
|
7D98
 
 | |
7DWO
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
5Z7H
 
 | |
5Z72
 
 | |
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
2H98
 
 | |
2H99
 
 | | Crystal structure of the effector binding domain of a BenM variant (R156H,T157S) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
2H9B
 
 | | Crystal structure of the effector binding domain of a BenM variant (BenM R156H/T157S) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
3HHG
 
 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
3ISP
 
 | | Crystal structure of ArgP from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator Rv1985c/MT2039 | | Authors: | Zhou, X, Lou, Z, Sheng, F, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2009-08-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ArgP from Mycobacterium tuberculosis Confirms Two Distinct Conformations of Full-length LysR Transcriptional Regulators and Reveals Its Function in DNA Binding and Transcriptional Regulation.
J.Mol.Biol., 2009
|
|
3FZJ
 
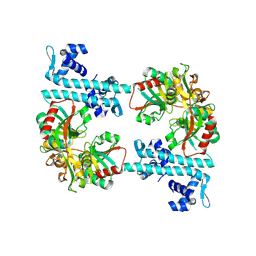 | | TsaR low resolution crystal structure, tetragonal form | | Descriptor: | LysR type regulator of tsaMBCD | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Dix, I, Kikhney, A.G, Svergun, D.I, Uson, I. | | Deposit date: | 2009-01-26 | | Release date: | 2010-01-26 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold.
Mol.Microbiol., 75, 2010
|
|
3FXU
 
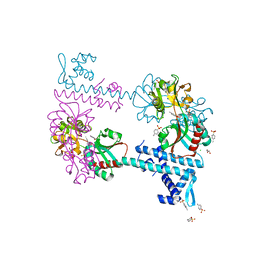 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3K1M
 
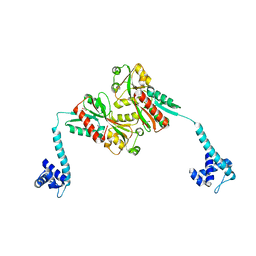 | | Crystal Structure of full-length BenM, R156H mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ruangprasert, A, Momany, C, Neidle, E.L, Craven, S.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Full-length BenM
To be Published
|
|
3K1P
 
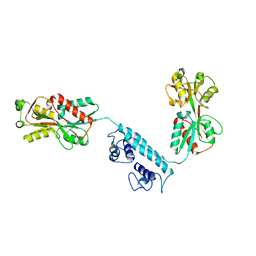 | |
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
