2GD6
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
1AJO
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1A7C
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR TYPE-1 IN COMPLEX WITH A PENTAPEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-ribopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAPEPTIDE, ... | | Authors: | Xue, Y, Inghardt, T, Sjolin, L, Deinum, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interfering with the inhibitory mechanism of serpins: crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactive-centre loop peptide
Structure, 6, 1998
|
|
1UH4
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1/malto-tridecaose complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
5Q03
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methylpropyl)thiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(2-methylpropyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methylpropyl)thiophen-2-yl]sulfonylurea
To be published
|
|
5Q01
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-[(3,4-dichloro-1-methyl-1H-pyrazol-5-yl)oxy]benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea
To be published
|
|
1IGR
 
 | | Type 1 Insulin-like growth factor receptor (DOMAINS 1-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, INSULIN-LIKE GROWTH FACTOR RECEPTOR 1, ... | | Authors: | Garrett, T.P.J, Mckern, N.M, Lou, M, Frenkel, M.J, Bentley, J.D, Lovrecz, G.O, Elleman, T.C, Cosgrove, L.J, Ward, C.W. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor.
Nature, 394, 1998
|
|
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2AH9
 
 | | Crystal Structure of Human M340H-Beta-1,4-Galactosyltransferase-I (M340H-B4Gal-T1) in Complex with Chitotriose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
3SKU
 
 | | Herpes simplex virus glycoprotein D bound to the human receptor nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D, ... | | Authors: | Di Giovine, P, Settembre, E.C, Bhargava, A.K, Luftig, M.A, Lou, H, Cohen, G.H, Eisenberg, R.J, Krummenacher, C, Carfi, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of herpes simplex virus glycoprotein d bound to the human receptor nectin-1.
Plos Pathog., 7, 2011
|
|
2O5R
 
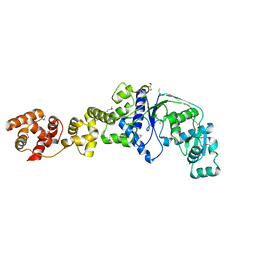 | |
3NWD
 
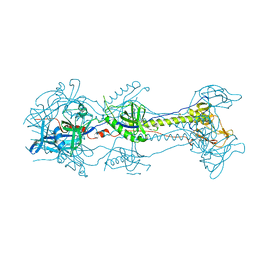 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8803 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
1CEB
 
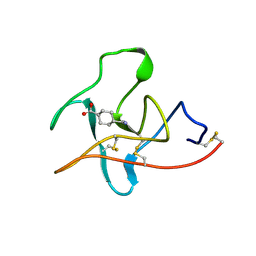 | |
5T4C
 
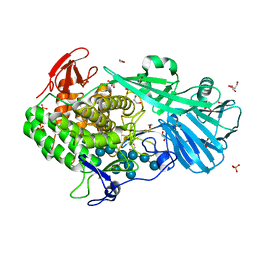 | | Crystal structure of BhGH81 mutant in complex with laminaro-hexaose | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, PHOSPHATE ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-08-29 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 81 Glycoside Hydrolase Implicates Its Recognition of beta-1,3-Glucan Quaternary Structure.
Structure, 25, 2017
|
|
2H5S
 
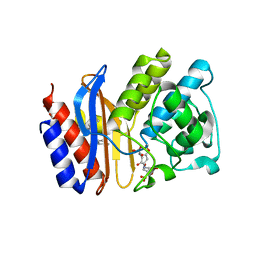 | | SA2-13 penam sulfone complexed to wt SHV-1 beta-lactamase | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, SHV-1 beta-lactamase | | Authors: | van den Akker, F, Padayatti, P.S. | | Deposit date: | 2006-05-27 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Rational Design of a beta-Lactamase Inhibitor Achieved via Stabilization of the trans-Enamine Intermediate: 1.28 A Crystal Structure of wt SHV-1 Complex with a Penam Sulfone.
J.Am.Chem.Soc., 128, 2006
|
|
2GT9
 
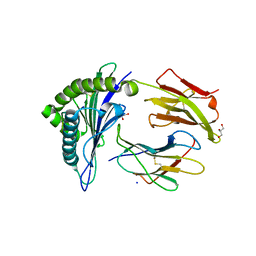 | |
1KKT
 
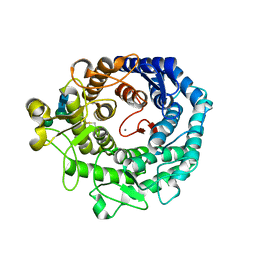 | | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannosyl-oligosaccharide alpha-1,2-mannosidase, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
2GQ1
 
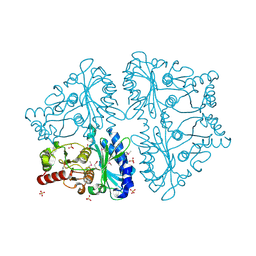 | | Crystal Structure of Recombinant Type I Fructose-1,6-bisphosphatase from Escherichia coli Complexed with Sulfate Ions | | Descriptor: | Fructose-1,6-bisphosphatase, SULFATE ION | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-04-19 | | Release date: | 2006-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Allosteric Activation Site in Escherichia coli Fructose-1,6-bisphosphatase.
J.Biol.Chem., 281, 2006
|
|
2MA9
 
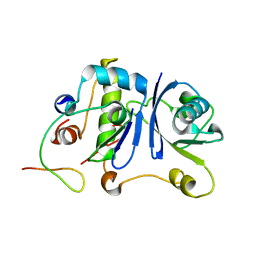 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
1ACY
 
 | |
1B0S
 
 | | BINDING OF AR-1-144, A TRI-IMIDAZOLE DNA MINOR GROOVE BINDER, TO CCGG SEQUENCE ANALYZED BY NMR SPECTROSCOPY | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, DNA (5'-D(*GP*AP*AP*CP*CP*GP*GP*TP*TP*C)-3') | | Authors: | Yang, X.-L, Kaenzig, C, Lee, M, Wang, A.H.-J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Binding of AR-1-144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy.
Eur.J.Biochem., 263, 1999
|
|
2HBO
 
 | |
2M1A
 
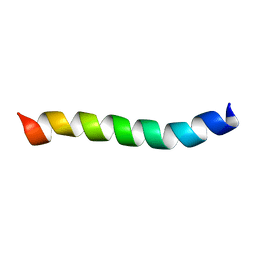 | | HIV-1 Rev ARM peptide (residues T34-R50) | | Descriptor: | HIV-1 Rev arginine-rich motif (ARM) | | Authors: | Casu, F, Duggan, B.M, Hennig, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Arginine-Rich RNA-Binding Motif of HIV-1 Rev Is Intrinsically Disordered and Folds upon RRE Binding.
Biophys.J., 105, 2013
|
|
2J9K
 
 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor MVT-101 | | Descriptor: | ACETATE ION, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
9PQ6
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | 1,2-ETHANEDIOL, 17-chloranyl-33-fluoranyl-5,13,14,22-tetramethyl-28-oxa-9-thia-5,6,12,13,24-pentazaheptacyclo[27.7.1.1^{4,7}.0^{11,15}.0^{16,21}.0^{20,24}.0^{30,35}]octatriaconta-1(36),4(38),6,11,14,16,18,20,22,29(37),30(35),31,33-tridecaene-23-carboxylic acid, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2025-07-22 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | In Pursuit of Best-in-Class MCL-1 Inhibitors: Discovery of Highly Potent 1,4-Indoyl Macrocycles with Favorable Physicochemical Properties.
J.Med.Chem., 68, 2025
|
|
