6T7O
 
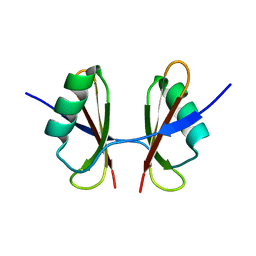 | | X-ray structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Fatkhullin, B.F, Khusainov, I.S, Ayupov, R.K, Gabdulkhakov, A.G, Tishchenko, S.V, Validov, S.Z, Yusupov, M.M. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60003626 Å) | | Cite: | Dimerization of long hibernation promoting factor from Staphylococcus aureus: Structural analysis and biochemical characterization.
J.Struct.Biol., 209, 2020
|
|
1CI1
 
 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5CCH
 
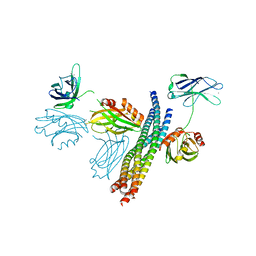 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
3WIP
 
 | | Crystal structure of acetylcholine bound to Ls-AChBP | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholine-binding protein, ... | | Authors: | Olsen, J.A, Balle, T, Gajhede, M, Ahring, P.K, Kastrup, J.S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of the neurotransmitter acetylcholine by an acetylcholine binding protein reveals determinants of binding to nicotinic acetylcholine receptors
Plos One, 9, 2014
|
|
4JPB
 
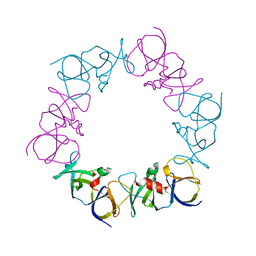 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
3HL4
 
 | | Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase with CDP-choline | | Descriptor: | Choline-phosphate cytidylyltransferase A, FORMIC ACID, GLYCEROL, ... | | Authors: | Lee, J, Paetzel, M, Cornell, R.B. | | Deposit date: | 2009-05-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a mammalian CTP: Phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyl-transferase fold
J.Biol.Chem., 284, 2009
|
|
6Q2U
 
 | | Structure of Cytochrome C4 from Pseudomonas aeruginosa | | Descriptor: | Cytochrome c4, HEME C | | Authors: | Carpenter, J.M, Zhong, F, Pletneva, E.V, Ragusa, M.J. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and redox properties of the diheme electron carrier cytochrome c4from Pseudomonas aeruginosa.
J.Inorg.Biochem., 203, 2019
|
|
8EBO
 
 | | Homopurine parallel G-quadruplex from human chromosome 7 stabilized by K+ ions | | Descriptor: | COBALT (III) ION, DNA (5'-D(*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*GP*A)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Chen, E.V, Yatsunyk, L.A. | | Deposit date: | 2022-08-31 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Homopurine guanine-rich sequences in complex with N-methyl mesoporphyrin IX form parallel G-quadruplex dimers and display a unique symmetry tetrad.
Bioorg.Med.Chem., 77, 2022
|
|
8EDP
 
 | | Crystal structure of a three-tetrad, parallel, and K+ stabilized homopurine G-quadruplex from human chromosome 7 | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Ye, M, Yatsunyk, L.A. | | Deposit date: | 2022-09-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Homopurine guanine-rich sequences in complex with N-methyl mesoporphyrin IX form parallel G-quadruplex dimers and display a unique symmetry tetrad.
Bioorg.Med.Chem., 77, 2022
|
|
4O5P
 
 | | Crystal structure of an uncharacterized protein from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein | | Authors: | Hu, H.D, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structure of the type VI secretion phospholipase effector Tle1 provides insight into its hydrolysis and membrane targeting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KF1
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 5 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
6QJH
 
 | | Cryo-EM structure of heparin-induced 2N4R tau snake filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
4OD4
 
 | |
8E55
 
 | |
4OFC
 
 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
8DUT
 
 | | Duplex-G-quadruplex-duplex (DGD) class_1 | | Descriptor: | Duplex-G-quadruplex-duplex (46-MER) | | Authors: | Monsen, R.C, Chua, E.Y.D, Hopkins, J.B, Chaires, J.B, Trent, J.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 angstrom resolution with cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
6WCU
 
 | | Crystal structure of coiled coil region of human septin 5 | | Descriptor: | Septin-5 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6WSM
 
 | | Crystal structure of coiled coil region of human septin 8 | | Descriptor: | SULFATE ION, Septin-8 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
5KGH
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KF2
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 8 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, Tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KF9
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGA
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
6QPW
 
 | | Structural basis of cohesin ring opening | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sister chromatid cohesion protein 1, ... | | Authors: | Panne, D, Muir, K.W, Li, Y, Weis, F. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Nat.Struct.Mol.Biol., 27, 2020
|
|
