5KB5
 
 | | Crystal structure of the adenosine kinase from Mus musculus in complex with adenosine and adenosine-diphosphate | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Adenosine kinase, ... | | Authors: | Oliveira, R.R, Neto, R.M, Polo, C.C, Tonoli, C.C.C, Murakami, M.T, Franchini, K.G. | | Deposit date: | 2016-06-02 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the adenosine kinase from Mus musculus in complex with adenosine and adenosine-diphosphate
To Be Published
|
|
5KC9
 
 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-1 (GluD1) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6D46
 
 | |
6D5A
 
 | |
4GHD
 
 | | Structure of Y257F variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.85 Ang resolution | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the role of tyrosine 257 of homoprotocatechuate 2,3-dioxygenase in substrate and oxygen activation.
Biochemistry, 51, 2012
|
|
5KGA
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
4D5E
 
 | | Crystal Structure of recombinant wildtype CDH | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-03 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4PXF
 
 | |
2G83
 
 | | Structure of activated G-alpha-i1 bound to a nucleotide-state-selective peptide: Minimal determinants for recognizing the active form of a G protein alpha subunit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Johnston, C.A, Ramer, J.K, Blaesius, R, Kuhlman, B, Arshavsky, V.Y, Siderovski, D.P. | | Deposit date: | 2006-03-01 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Minimal Determinants for Binding Activated Galpha from the Structure of a Galpha(i1)-Peptide Dimer.
Biochemistry, 45, 2006
|
|
5KLV
 
 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
6DCX
 
 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
5KM5
 
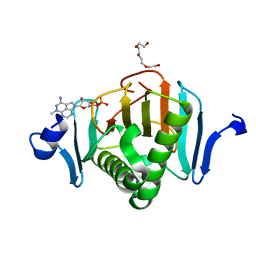 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) triciribine 5'-monoposphate catalytic product complex | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, CHLORIDE ION, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KZU
 
 | | Crystal structure of an acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole | | Descriptor: | 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Wu, J, Talley, I.T, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole
To Be Published
|
|
4GHF
 
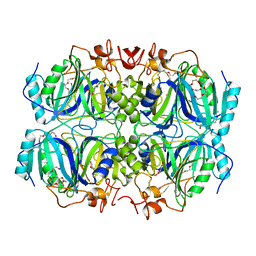 | | Structure of Y257F variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-Nitrocatechol and dioxygen at 1.67 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for the role of tyrosine 257 of homoprotocatechuate 2,3-dioxygenase in substrate and oxygen activation.
Biochemistry, 51, 2012
|
|
5KM9
 
 | |
5L88
 
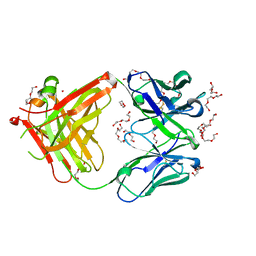 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L9D
 
 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSYLATED, CRYSTAL FORM I, parsimonious model | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-10 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2GA3
 
 | |
4HNA
 
 | |
4QGP
 
 | |
4QHB
 
 | |
4CW3
 
 | | Crystal structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-metyl uric acid (X-ray dose, 665 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, 9-METHYL URIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
2YHK
 
 | | D214A mutant of tyrosine phenol-lyase from Citrobacter freundii | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Citrobacter Freundii Asp214Ala Tyrosine Phenol-Lyase Reveals that Asp214 is Critical for Maintaining a Strain in the Internal Aldimine
Croatica Chemica Acta, 85, 2012
|
|
