5VJ6
 
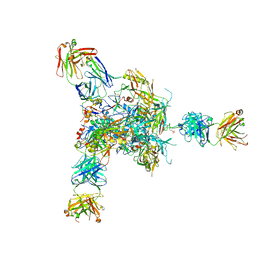 | |
7LIF
 
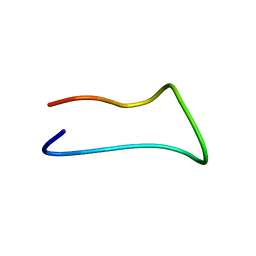 | |
5W3V
 
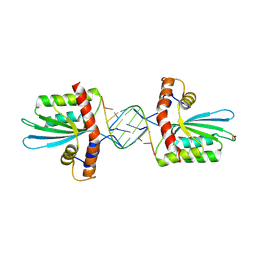 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
7LVF
 
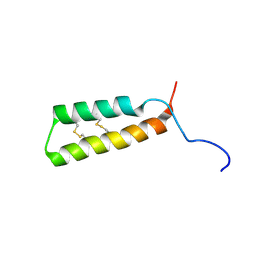 | |
5IOH
 
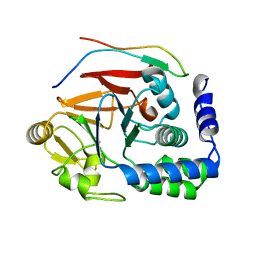 | | RepoMan-PP1a (protein phosphatase 1, alpha isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
7LQT
 
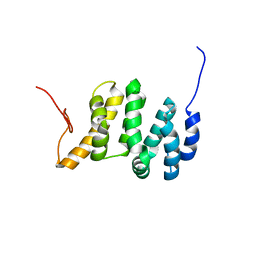 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
5IOP
 
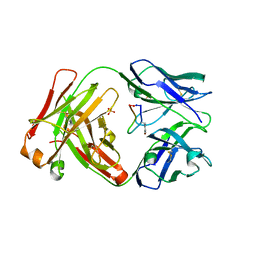 | |
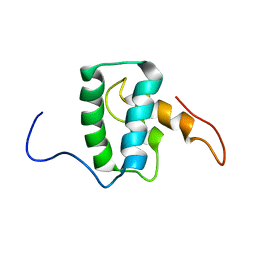
7M1W
 
 | |
5IP5
 
 | |
7M2A
 
 | |
7M77
 
 | |
5VDE
 
 | |
7LSY
 
 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
7MSF
 
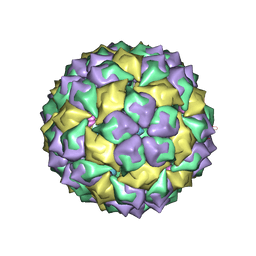 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
5IPR
 
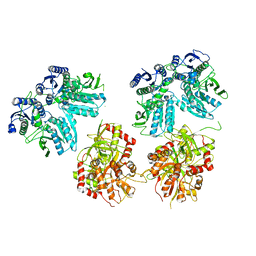 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 3 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IQ1
 
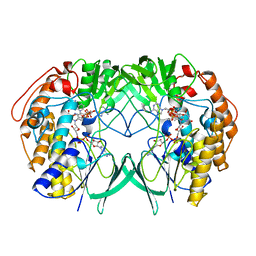 | | Crystal structure of RnTmm mutant Y207S | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5VNN
 
 | |
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
5IEY
 
 | | Crystal structure of a CDK inhibitor bound to CDK2 | | Descriptor: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
5IFI
 
 | | CRYSTAL STRUCTURE OF ACETYL-COA SYNTHETASE IN COMPLEX WITH ADENOSINE-5'-PROPYLPHOSPHATE FROM CRYPTOCOCCUS NEOFORMANS H99 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), SSGCID, Fox III, D, Edwards, T.E, Lorimer, D.D, Mutz, M.W. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-16 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and evaluation of acyl-AMP phosphate isosteres as inhibitors of fungal acetyl CoA synthetase.
Bioorg.Med.Chem.Lett., 2025
|
|
5VEJ
 
 | |
5IQH
 
 | |
5IRF
 
 | |
