3C32
 
 | |
4KFM
 
 | |
3E83
 
 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
7C75
 
 | | Crystal structure of yak lactoperoxidase with partially coordinated Na ion in the distal heme cavity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Rani, C, Ahmad, N, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
2RGI
 
 | |
3NPS
 
 | |
7DJC
 
 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
2ITC
 
 | | Potassium Channel KcsA-Fab complex in Sodium Chloride | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, SODIUM ION, ... | | Authors: | Lockless, S.W, Zhou, M, MacKinnon, R. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Thermodynamic Properties of Selective Ion Binding in a K(+) Channel.
Plos Biol., 5, 2007
|
|
8CTW
 
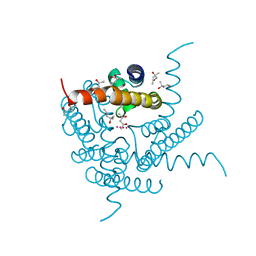 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Na+,Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
3FSY
 
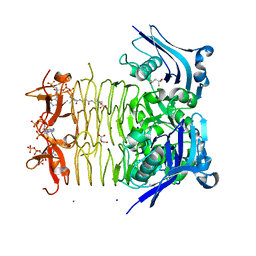 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c;DapD) in complex with succinyl-CoA from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
3FSX
 
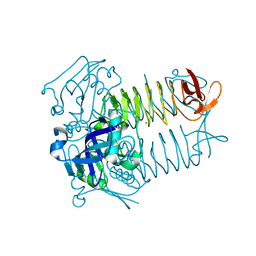 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c; DapD) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
3NT8
 
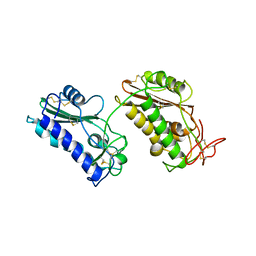 | | Crystal Structure of Na-ASP-1 | | Descriptor: | Ancylostoma secreted protein 1 | | Authors: | Asojo, O.A. | | Deposit date: | 2010-07-02 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a two-CAP-domain protein from the human hookworm parasite Necator americanus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7EVI
 
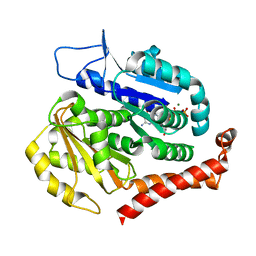 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 79% GTP, 21% GDP, Na+, Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVE
 
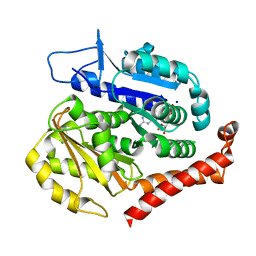 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 100% GDP and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, Tubulin-like protein | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVC
 
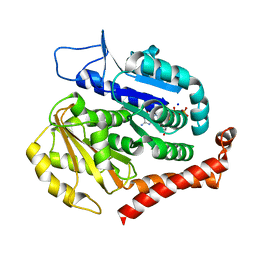 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 60% GTP/40% GDP and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVL
 
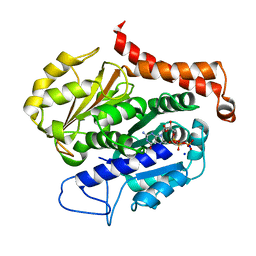 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 64% GTP/36% GDP and 2 Na+ in a small unit cell | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVD
 
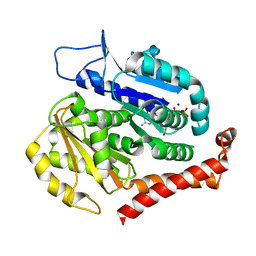 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 53% GTP/47% and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVK
 
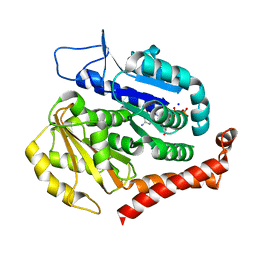 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 78% GTP, 22% GDP, Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C, Tran, L.T. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
7EVB
 
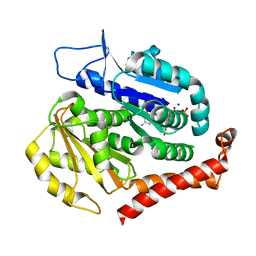 | | Odinarchaeota tubulin (OdinTubulin) H393D mutant, in a protofilament arrangement, bound to 77% GTP/23% and 2 Na+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, SODIUM ION, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution.
Sci Adv, 8, 2022
|
|
4DL8
 
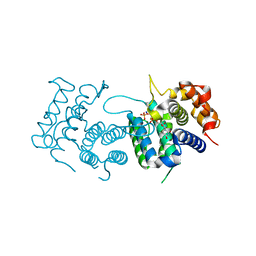 | | Crystal structure of Trypanosoma brucei dUTPase with dUMP, planar [AlF3-OPO3] transition state analogue, Mg2+, and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ALUMINUM FLUORIDE, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
3VOU
 
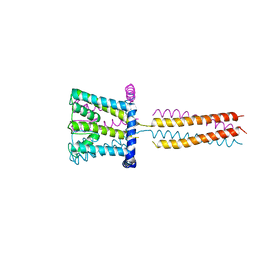 | | The crystal structure of NaK-NavSulP chimera channel | | Descriptor: | COBALT (II) ION, Ion transport 2 domain protein, Voltage-gated sodium channel, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate
Nat Commun, 3, 2012
|
|
4DII
 
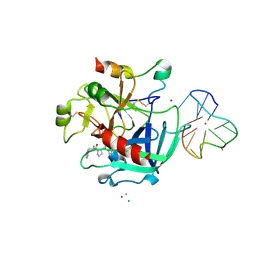 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of potassium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
3B8E
 
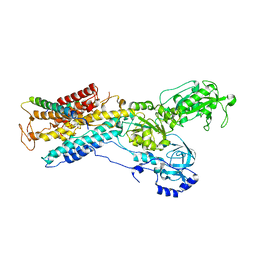 | | Crystal structure of the sodium-potassium pump | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, ... | | Authors: | Morth, J.P, Pedersen, P.B, Toustrup-Jensen, M.S, Soerensen, T.L.M, Petersen, J, Andersen, J.P, Vilsen, B, Nissen, P. | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-potassium pump.
Nature, 450, 2007
|
|
4ED3
 
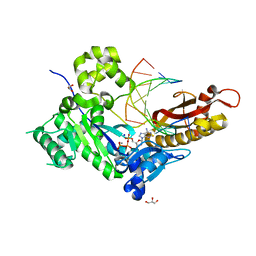 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 7.5 (Na+ HEPES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4D79
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
