3NZS
 
 | |
3O0G
 
 | | Crystal Structure of Cdk5:p25 in complex with an ATP analogue | | Descriptor: | Cell division protein kinase 5, Cyclin-dependent kinase 5 activator 1, {4-amino-2-[(4-chlorophenyl)amino]-1,3-thiazol-5-yl}(3-nitrophenyl)methanone | | Authors: | Mapelli, M. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation
Chem.Biol., 12, 2005
|
|
3O2L
 
 | |
3O3B
 
 | | Human Class I MHC HLA-A2 in complex with the Peptidomimetic ELA-1.1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2010-07-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with Melan-A/MART-1(26(27L)-35) peptidomimetics reveal conformational heterogeneity and highlight degeneracy of T cell recognition.
J.Med.Chem., 53, 2010
|
|
3NPG
 
 | |
3NT5
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9006 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTO
 
 | |
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
3NO3
 
 | |
3NOK
 
 | | Crystal structure of Myxococcus xanthus Glutaminyl Cyclase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
2BCX
 
 | | Crystal structure of calmodulin in complex with a ryanodine receptor peptide | | Descriptor: | CALCIUM ION, Calmodulin, Ryanodine receptor 1 | | Authors: | Maximciuc, A.A, Shamoo, Y, MacKenzie, K.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode.
Structure, 14, 2006
|
|
3NP6
 
 | | The crystal structure of Berberine bound to DNA d(CGTACG) | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', BERBERINE, CALCIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2010-06-28 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG)
Chem.Commun.(Camb.), 47, 2011
|
|
3NPD
 
 | |
3NT2
 
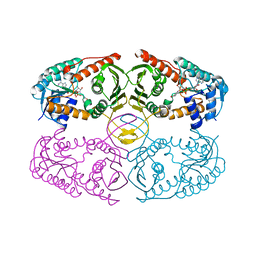 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3003 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3O47
 
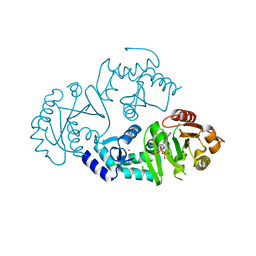 | | Crystal structure of ARFGAP1-ARF1 fusion protein | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Tong, Y, Nedyalkova, L, Tempel, W, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ARFGAP1-ARF1 fusion protein
to be published
|
|
3NN8
 
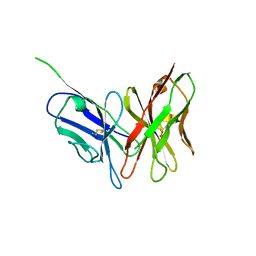 | |
2IT9
 
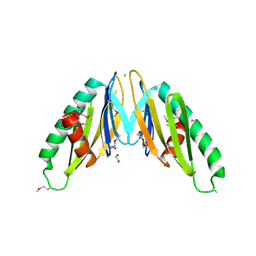 | |
3NVL
 
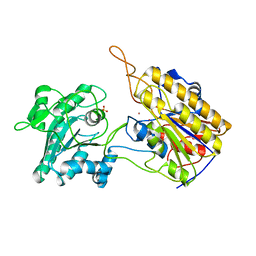 | | Crystal Structure of Phosphoglycerate Mutase from Trypanosoma brucei | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, COBALT (II) ION, SULFATE ION | | Authors: | Mercaldi, G.F, Pereira, H.M, Cordeiro, A.T, Andricopulo, A.D, Thiemann, O.H. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphoglycerate mutase from Trypanosoma brucei.
Febs J., 279, 2012
|
|
3NYO
 
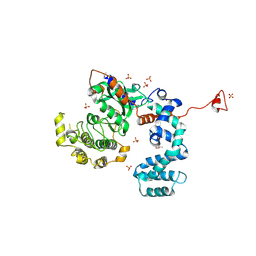 | | Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with AMP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE MONOPHOSPHATE, G protein-coupled receptor kinase 6, ... | | Authors: | Tesmer, J.J.G, Singh, P. | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Molecular basis for activation of G protein-coupled receptor kinases.
Embo J., 29, 2010
|
|
3O3L
 
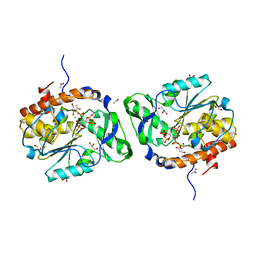 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,3,4,5)tetrakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of substrate binding in PTPLPs
To be Published
|
|
3O46
 
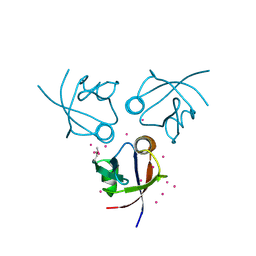 | | Crystal structure of the PDZ domain of MPP7 | | Descriptor: | MAGUK p55 subfamily member 7, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Zhong, N, Guan, X, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the PDZ domain of MPP7
TO BE PUBLISHED
|
|
2ICG
 
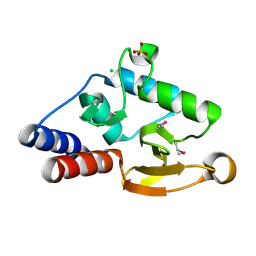 | |
3O6O
 
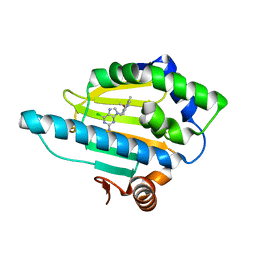 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 83, PENTAETHYLENE GLYCOL | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Li, Y, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P, Fairlamb, A.H, Ferguson, M.A.J, Thompson, S, MacKenzie, C, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
3OA3
 
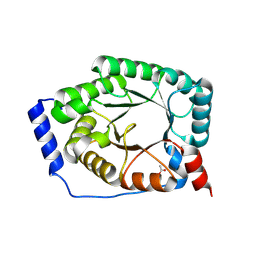 | |
3O8Z
 
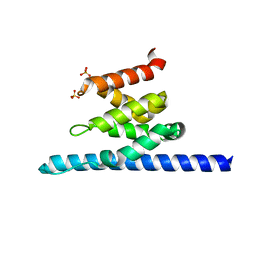 | | Crystal structure of Spn1 (Iws1) core domain | | Descriptor: | SULFATE ION, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
