8Z5G
 
 | |
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
9J9C
 
 | |
9FJU
 
 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9C7V
 
 | | Structure of the human BOS:human EMC complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
5AXH
 
 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
9C4G
 
 | |
8ZBQ
 
 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
9BCG
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 67, 2024
|
|
9ATO
 
 | | XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3C, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of XBB.1.5 spike/Nanosota-3C complex
To Be Published
|
|
9FFD
 
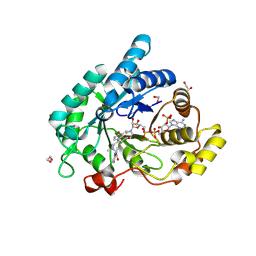 | | STRUCTURE OF ALDO-KETO REDUCTASE 1C3 (AKR1C3) IN COMPLEX WITH AN INHIBITOR MEDS765 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldo-keto reductase family 1 member C3, ... | | Authors: | Frydenvang, K, Hussain, S, Mirza, O.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | AI Based Discovery of a New AKR1C3 Inhibitor for Anticancer Applications.
Acs Med.Chem.Lett., 15, 2024
|
|
8YS2
 
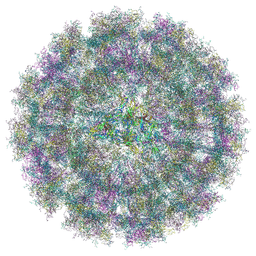 | | Overall structure of Eastern Equine Encephalitis virus VLP in complex with the receptor VLDLR LA1-2 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2024-03-22 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
8YS4
 
 | | Overall structure of Eastern Equine Encephalitis virus VLP in complex with the receptor VLDLR LA3-5 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2024-03-22 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
9FL0
 
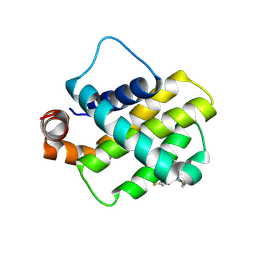 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[(4-chlorophenyl)methyl]-~{N}-(4-fluorophenyl)prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9EUO
 
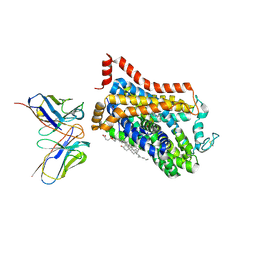 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
8ZKC
 
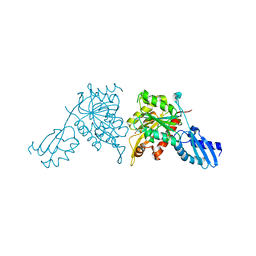 | | iron-sulfur cluster transfer protein ApbC | | Descriptor: | GLYCEROL, Iron-sulfur cluster carrier protein, MAGNESIUM ION | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the iron-sulfur cluster transfer protein ApbC from Escherichia coli.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
9FKY
 
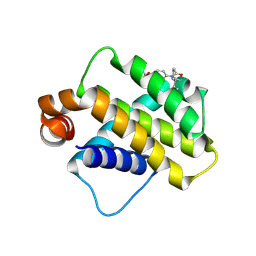 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(1~{S})-1-[3-cyano-4-(trifluoromethyl)phenyl]ethyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9J2N
 
 | | Cryo-EM structure of the human glucose transporter, GLUT7 in outward-facing open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 2, facilitated glucose transporter member 7 | | Authors: | Lee, S.S, Kim, S, Jin, M.S. | | Deposit date: | 2024-08-07 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human glucose transporter GLUT7.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
8ZPB
 
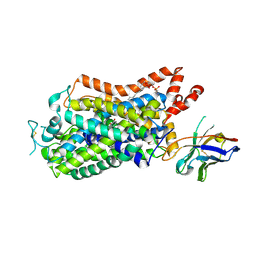 | |
5ANN
 
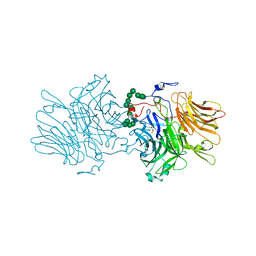 | | Structure of fructofuranosidase from Xanthophyllomyces dendrorhous | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-FRUCTOFURANOSIDASE, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-09-07 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
9GAO
 
 | | Crystal structure of CRBNmidi in complex with 2-(4-(2,6-dioxopiperidin-3-yl)phenoxy)-N-methylacetamide | | Descriptor: | 2-[4-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]phenoxy]-~{N}-methyl-ethanamide, Protein cereblon, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
To Be Published
|
|
9EUP
 
 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
9IIW
 
 | |
9GII
 
 | | Jumonji domain-containing protein 2A with crystallization epitope mutation R913A | | Descriptor: | GLYCEROL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
8ZMF
 
 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
