5ZWK
 
 | | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with fructose-1,6-bisphophate and AMP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Zeyuan, G, Junjie, Y, Ping, Y, Jian, W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Location of FBPase catalytic metal binding site: a combined experimental and theoretical study
To Be Published
|
|
7BXO
 
 | | Crystal structure of the toxin-antitoxin with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Toxin-antitoxin system antidote Mnt family, ... | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
5L95
 
 | | Crystal structure of human UBA5 in complex with UFM1 and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-fold modifier 1, Ubiquitin-like modifier-activating enzyme 5, ... | | Authors: | Oweis, W, Padala, P, Wiener, R. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-Binding Mechanism of Ubiquitin-like Protein Activation Revealed by a UBA5-UFM1 Complex.
Cell Rep, 16, 2016
|
|
5JDA
 
 | | Bacillus cereus CotH kinase plus Mg2+/AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Phosphorylation of spore coat proteins by a family of atypical protein kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
1UXP
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1SZM
 
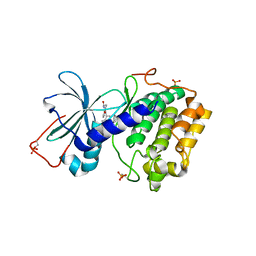 | | DUAL BINDING MODE OF BISINDOLYLMALEIMIDE 2 TO PROTEIN KINASE A (PKA) | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Gassel, M, Breitenlechner, C.B, Koenig, N, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The protein kinase C inhibitor bisindolyl maleimide 2 binds with reversed orientations to different conformations of protein kinase a.
J.Biol.Chem., 279, 2004
|
|
3LLA
 
 | |
6YJ9
 
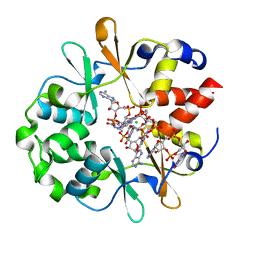 | | DarB in complex with 3'3'cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CBS domain-containing protein YkuL, CHLORIDE ION | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DarB from B. subtilis
To Be Published
|
|
8IMF
 
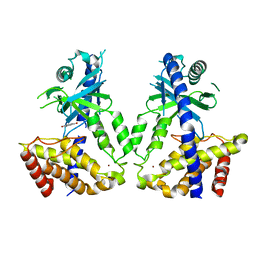 | | Human cGAS catalytic domain bound with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IME
 
 | | Human cGAS catalytic domain bound with baicalin | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
8IMG
 
 | | Human cGAS catalytic domain bound with C20 | | Descriptor: | 2-[2-hydroxy-2-oxoethyl-[3-(7-methoxy-4-methyl-2-oxidanylidene-chromen-3-yl)propanoyl]amino]ethanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Li, J.M, Xu, Y.C. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel cGAS inhibitors based on natural flavonoids.
Bioorg.Chem., 140, 2023
|
|
1M0W
 
 | | Yeast Glutathione Synthase Bound to gamma-glutamyl-cysteine, AMP-PNP and 2 Magnesium Ions | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-14 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Conformational Changes in the Catalytic Cycle of Glutathione Synthase
Structure, 10, 2002
|
|
1FY2
 
 | | Aspartyl Dipeptidase | | Descriptor: | ASPARTYL DIPEPTIDASE, CADMIUM ION | | Authors: | Hakansson, K, Wang, A.H.-J, Miller, C.G. | | Deposit date: | 2000-09-28 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FYE
 
 | |
3IM3
 
 | | Crystal structure of PKA RI alpha dimerization/docking domain | | Descriptor: | FORMIC ACID, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Sarma, G.N, Kinderman, F.S, Kim, C, von Daake, S, Taylor, S.S. | | Deposit date: | 2009-08-09 | | Release date: | 2010-02-02 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of D-AKAP2:PKA RI Complex: Insights into AKAP Specificity and Selectivity
Structure, 18, 2010
|
|
8JA2
 
 | |
8JA1
 
 | |
2QYK
 
 | | Crystal structure of PDE4A10 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, Cyclic AMP-specific phosphodiesterase HSPDE4A10, MAGNESIUM ION, ... | | Authors: | Wang, H, Peng, M, Chen, Y, Geng, J, Robinson, H, Houslay, M. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
6VT1
 
 | | Naegleria gruberi RNA ligase D172A mutant apo | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
2JGD
 
 | | E. COLI 2-oxoglutarate dehydrogenase (E1o) | | Descriptor: | 2-OXOGLUTARATE DEHYDROGENASE E1 COMPONENT, ADENOSINE MONOPHOSPHATE | | Authors: | Frank, R.A.W, Price, A.J, Northrop, F.D, Perham, R.N, Luisi, B.F. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
J.Mol.Biol., 368, 2007
|
|
3SP1
 
 | |
5VEO
 
 | | Murine ectonucleotide pyrophosphatase / phosphodiesterase 5 (ENPP5, NPP5), inactive (T72A), in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gorelik, A, Randriamihaja, A, Illes, K, Nagar, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A key tyrosine substitution restricts nucleotide hydrolysis by the ectoenzyme NPP5.
FEBS J., 284, 2017
|
|
1TBB
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Rolipram | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ROLIPRAM, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
2VNI
 
 | |
