4UCU
 
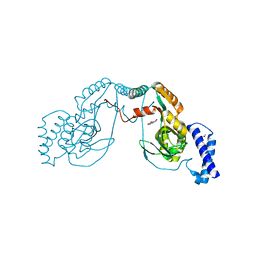 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4LMZ
 
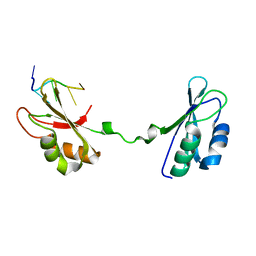 | |
6P1O
 
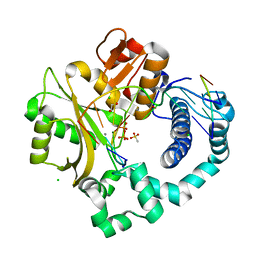 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated dAMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
3K2E
 
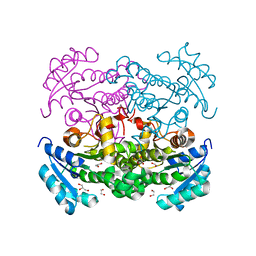 | |
5JPC
 
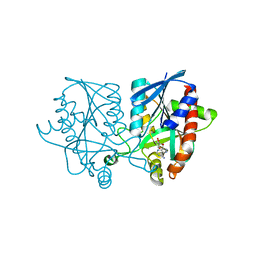 | | Joint X-ray/neutron structure of MTAN complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1CN4
 
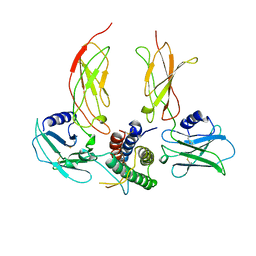 | |
5JV0
 
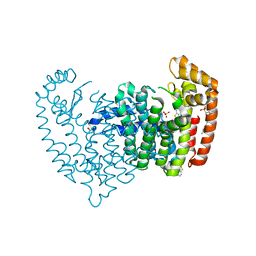 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-038 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-2-(3-fluorophenyl)-1-{[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
6PAC
 
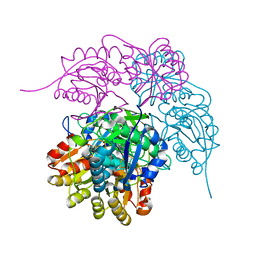 | |
3O1F
 
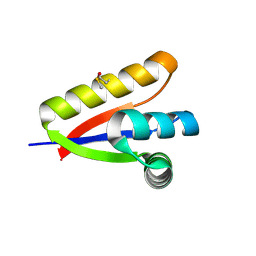 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | Descriptor: | ATP-dependent Clp protease adapter protein clpS | | Authors: | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-27 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
6U18
 
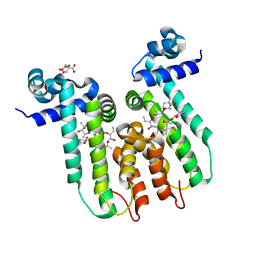 | | Directed evolution of a biosensor selective for the macrolide antibiotic clarithromycin | | Descriptor: | CITRATE ANION, CLARITHROMYCIN, Erythromycin resistance repressor protein | | Authors: | Li, Y, Reed, M, Wright, H.T, Cropp, T.A, Williams, G. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Genetically Encoded Biosensors for Reporting the Methyltransferase-Dependent Biosynthesis of Semisynthetic Macrolide Antibiotics.
Acs Synth Biol, 2021
|
|
6PJX
 
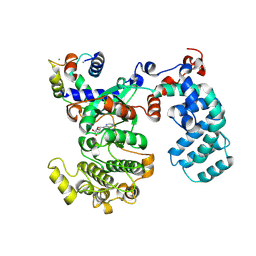 | | Crystal Structure of G Protein-Coupled Receptor Kinase 5 (GRK5) in Complex with Calmodulin (CaM) | | Descriptor: | CALCIUM ION, Calmodulin, G protein-coupled receptor kinase 5, ... | | Authors: | Bhardwaj, A, Komolov, K.E, Sulon, S, Benovic, J.L. | | Deposit date: | 2019-06-28 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a GRK5-Calmodulin Complex Reveals Molecular Mechanism of GRK Activation and Substrate Targeting.
Mol.Cell, 81, 2021
|
|
4LHO
 
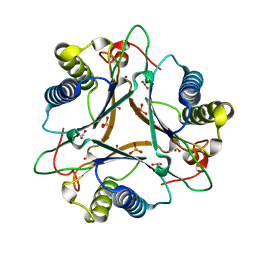 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4LKT
 
 | |
3JS5
 
 | |
5JX4
 
 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
5KBL
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110Q Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
8AIP
 
 | | Crystal Structure of Two-domain bacterial laccase from the actinobacterium Streptomyces carpinensis VKM Ac-1300 | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, Two-Domain Laccase | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Trubitsina, L, Trubitsin, I, Leontievsky, A, Lisov, A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Novel Two-Domain Laccase with Middle Redox Potential: Physicochemical and Structural Properties.
Biochemistry Mosc., 88, 2023
|
|
3SO0
 
 | |
5KEG
 
 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | Authors: | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | Deposit date: | 2016-06-09 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
3SDW
 
 | |
3O7M
 
 | | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor'
TO BE PUBLISHED
|
|
5JQ5
 
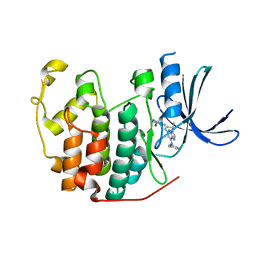 | | Crystal structure of CDK2 in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
5JQ8
 
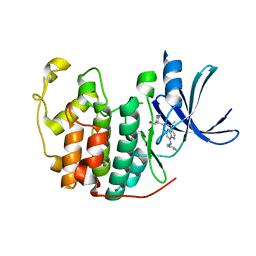 | | Crystal structure of CDK2 in complex with inhibitor ICEC0943 | | Descriptor: | (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
4LPW
 
 | |
