8U5K
 
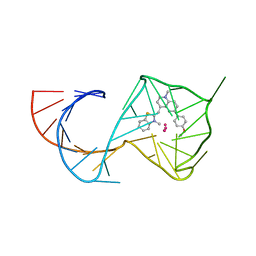 | |
8U5T
 
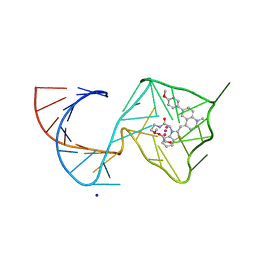 | | Structure of Mango II variant aptamer bound to T01-6A-B | | Descriptor: | 3-{2,16-dioxo-20-[(3aS,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-6A-B
To Be Published
|
|
8U5J
 
 | | Structure of Mango III variant aptamer bound to T01-07M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, DIMETHYL SULFOXIDE, Mango III variant, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mango III variant aptamer bound to T01-07M-B
To Be Published
|
|
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | Descriptor: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-05-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
190D
 
 | |
8YAX
 
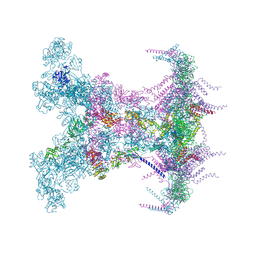 | | SARS-CoV-2 DMV nsp3-4 pore complex (full-pore) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-10 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB7
 
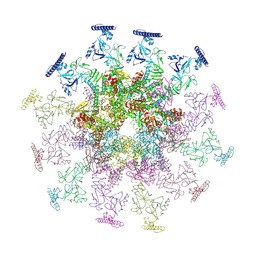 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C3 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB5
 
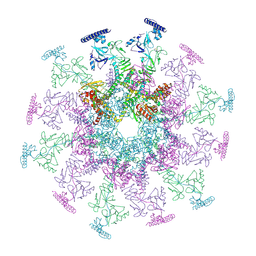 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8OST
 
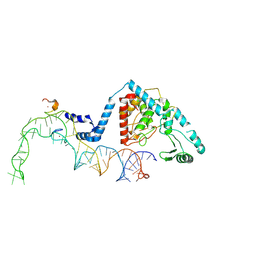 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPS
 
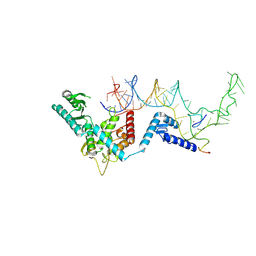 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 1 | | Descriptor: | Protein lin-28 homolog A, RNA (71-MER) Let7g, Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPT
 
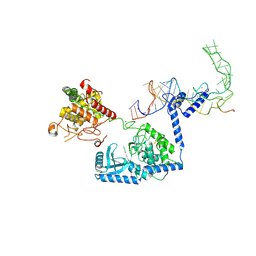 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | | Descriptor: | Protein lin-28 homolog A, RNA (53-MER), Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PO8
 
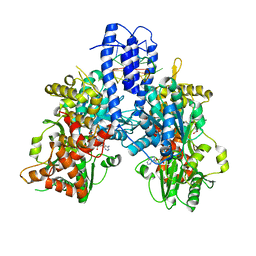 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO6
 
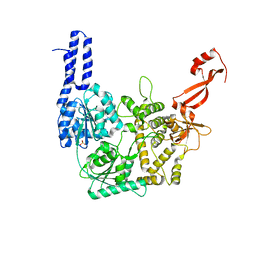 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO7
 
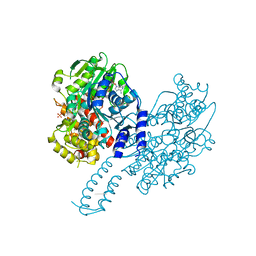 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PYW
 
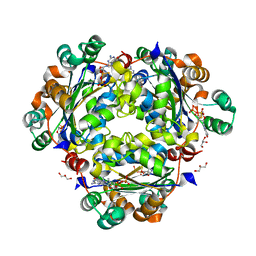 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8PIE
 
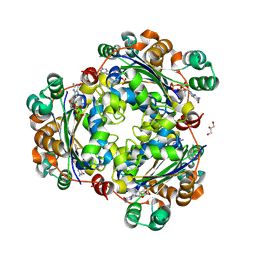 | | Crystal structure of the human nucleoside diphosphate kinase B domain in complex with the product AT-8500 formed by catalysis of compound AT-9010 | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Feracci, M, Chazot, A, Ferron, F, Alvarez, K, Canard, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
7WU8
 
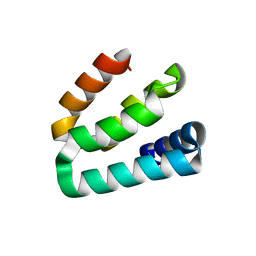 | |
2RSK
 
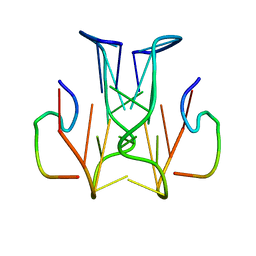 | | RNA aptamer against prion protein in complex with the partial binding peptide | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | Authors: | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
6HEG
 
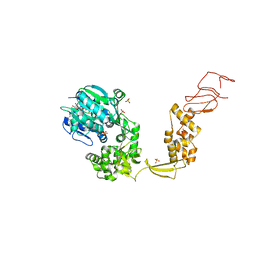 | | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpB, PHOSPHATE ION, ... | | Authors: | Xin, B.G, Chen, W.F, Rety, S, Dai, Y.X, Xi, X.G. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
7AAO
 
 | |
7AAF
 
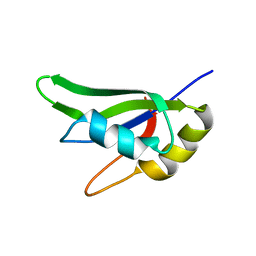 | |
1HD1
 
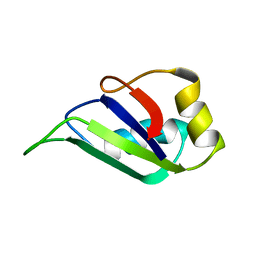 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
1HD0
 
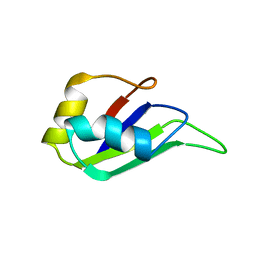 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
5FHD
 
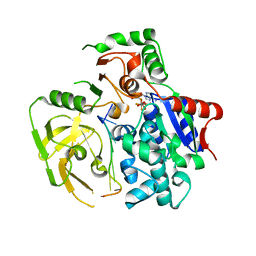 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FTB
 
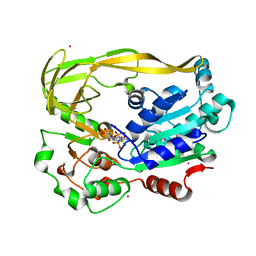 | | Crystal structure of Pif1 helicase from Bacteroides in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
