3JYS
 
 | |
1CRL
 
 | |
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2RF8
 
 | |
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2ZWR
 
 | | Crystal structure of TTHA1623 from thermus thermophilus HB8 | | Descriptor: | Metallo-beta-lactamase superfamily protein, ZINC ION | | Authors: | Yamamura, A, Okada, A, Kameda, Y, Ohtsuka, J, Nakagawa, N, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of TTHA1623, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZZI
 
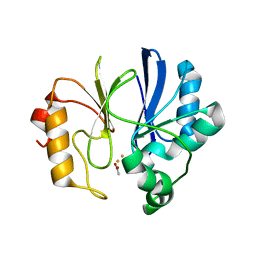 | | Crystal structure of TTHA1623 in a di-iron-bound form | | Descriptor: | ACETATE ION, FE (III) ION, Metallo-beta-lactamase superfamily protein | | Authors: | Yamamura, A, Okada, A, Kameda, Y, Ohtsuka, J, Nakagawa, N, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2009-02-16 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of TTHA1623, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6GV1
 
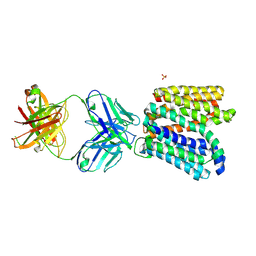 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
5J7R
 
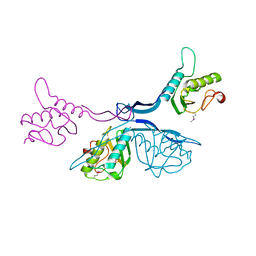 | | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-20 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To Be Published
|
|
3BUU
 
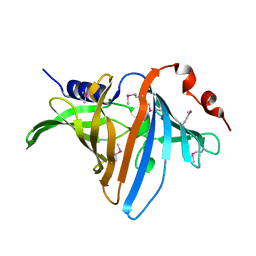 | |
3CCG
 
 | |
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
1BFJ
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1BFI
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, 30 STRUCTURES | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
8OYE
 
 | | Clostridium perfringens chitinase CP4_3455 E196Q with chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitodextrinase, DIMETHYL SULFOXIDE | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-05-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinase CP4_3455 E196Q with chitin
To Be Published
|
|
8OTB
 
 | |
7AFS
 
 | | The structure of Artemis variant D37A | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
2J0V
 
 | | The crystal structure of Arabidopsis thaliana RAC7-ROP9: the first RAS superfamily GTPase from the plant kingdom | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAC-LIKE GTP-BINDING PROTEIN ARAC7 | | Authors: | Sormo, C.G, Leiros, I, Brembu, T, Winge, P, Os, V, Bones, A.M. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Crystal Structure of Arabidopsis Thaliana Rac7/Rop9: The First Ras Superfamily Gtpase from the Plant Kingdom.
Phytochemistry, 67, 2006
|
|
7APV
 
 | | Structure of Artemis/DCLRE1C/SNM1C in complex with Ceftriaxone | | Descriptor: | 1,2-ETHANEDIOL, Ceftriaxone, NICKEL (II) ION, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AF1
 
 | | The structure of Artemis/SNM1C/DCLRE1C with 2 Zinc ions | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AFU
 
 | | The structure of Artemis variant H33A | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
1YMQ
 
 | |
8OSE
 
 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C
To Be Published
|
|
2XB1
 
 | | Crystal structure of the human Pygo2 PHD finger in complex with the B9L HD1 domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYGOPUS HOMOLOG 2, ... | | Authors: | Miller, T.C, Rutherford, T.J, Johnson, C.M, Fiedler, M, Bienz, M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Remodeling of the Histone H3 Binding Pocket in the Pygo2 Phd Finger Triggered by its Binding to the B9L/Bcl9 Co-Factor.
J.Mol.Biol., 401, 2010
|
|
