6NLW
 
 | | The crystal structure of class D carbapenem-hydrolyzing beta-lactamase BlaA from Shewanella oneidensis MR-1 | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-09 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of class D carbapenem-hydrolyzing beta-lactamase BlaA from Shewanella oneidensis MR-1
To Be Published
|
|
4QTY
 
 | | Caspase-3 E190A | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, AZIDE ION, Caspase-3, ... | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QU5
 
 | | Caspase-3 T140V | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | Descriptor: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
2JQ4
 
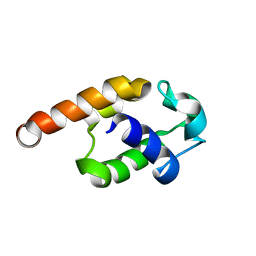 | | Complete resonance assignments and solution structure calculation of ATC2521 (NESG ID: AtT6) from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2571 | | Authors: | Srisailam, S, Lemak, A, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a protein (ATC2521)of unknown function from Agrobacterium tumefaciens
To be Published
|
|
5WKJ
 
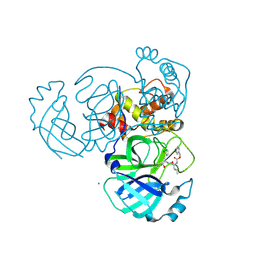 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, CALCIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKK
 
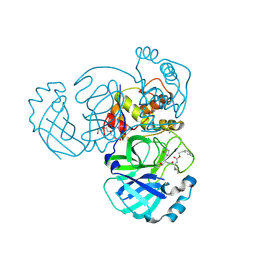 | | 1.55 A resolution structure of MERS 3CL protease in complex with inhibitor GC813 | | Descriptor: | (1R,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKM
 
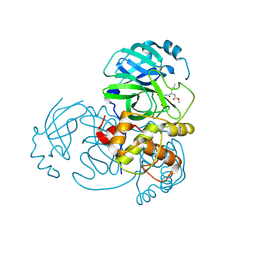 | | 2.25 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 21 | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
4H1Z
 
 | | Crystal structure of putative isomerase from Sinorhizobium meliloti, open loop conformation (target EFI-502104) | | Descriptor: | CHLORIDE ION, Enolase Q92Zs5, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Crystal Structure of Enolase Q92Zs5 (Target EFI-502104) from Sinorhizobium meliloti
To be Published
|
|
3DWD
 
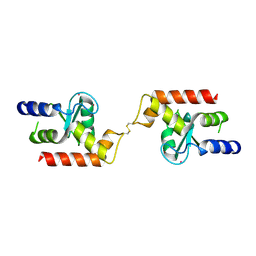 | | Crystal structure of the ArfGAP domain of human ARFGAP1 | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ArfGAP domain of human ARFGAP1
To be Published
|
|
2OZA
 
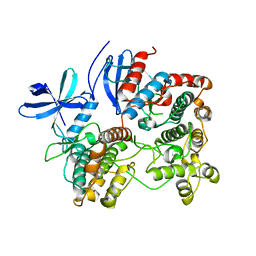 | | Structure of p38alpha complex | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | White, A, Pargellis, C.A, Studts, J.M, Werneburg, B.G, Farmer II, B.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of MAPK-activated protein kinase 2:p38 assembly
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5IKN
 
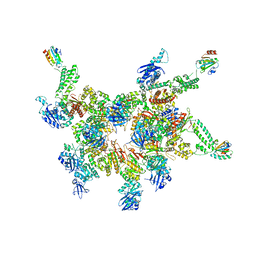 | |
6BXI
 
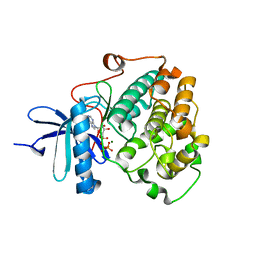 | | X-ray crystal structure of NDR1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | Authors: | Xiong, S, Sicheri, F. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
1S1R
 
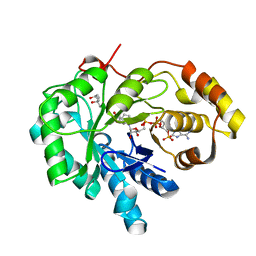 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
3ZFX
 
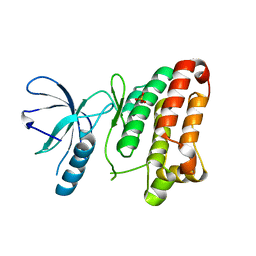 | | Crystal structure of EphB1 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 1, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
7M94
 
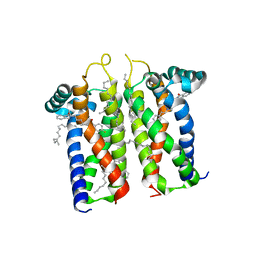 | | Bovine sigma-2 receptor bound to Roluperidone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Roluperidone, Sigma intracellular receptor 2 | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
6AUI
 
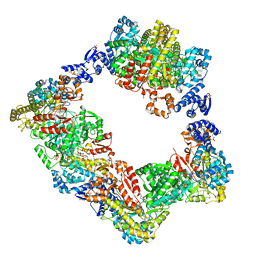 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
5C9X
 
 | |
6RGS
 
 | |
7K90
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | Authors: | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
5CBB
 
 | |
3ZWU
 
 | | Pseudomonas fluorescens PhoX in complex with vanadate, a transition state analogue | | Descriptor: | ALKALINE PHOSPHATASE PHOX, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
6PGO
 
 | | Crystal structure of human KRAS G12C covalently bound to a phthalazine inhibitor | | Descriptor: | 1-{4-[7-chloro-6-(2-fluoro-6-hydroxyphenyl)-4-phenylphthalazin-1-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Covalent Inhibitor of KRASG12C(AMG 510) for the Treatment of Solid Tumors.
J.Med.Chem., 63, 2020
|
|
1S67
 
 | | Crystal structure of heme domain of direct oxygen sensor from E. coli | | Descriptor: | Hypothetical protein yddU, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, H.J, Suquet, C, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH)
Biochemistry, 43, 2004
|
|
4R6B
 
 | | Rational Design of Enhanced Photoresistance in a Photoswitchable Fluorescent Protein | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Duan, C, Adam, V, Byrdin, M, Bourgeois, D. | | Deposit date: | 2014-08-23 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of enhanced photoresistance in a photoswitchable fluorescent protein.
Methods Appl Fluoresc, 3, 2015
|
|
