4MGH
 
 | | Importance of Hydrophobic Cavities in Allosteric Regulation of Formylglycinamide Synthetase: Insight from Xenon Trapping and Statistical Coupling Analysis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-08-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
3C0K
 
 | |
4M0F
 
 | | Structure of human acetylcholinesterase in complex with territrem B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Gary, E.N, Shiomi, K, Rosenberry, T.L. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structures of human acetylcholinesterase bound to dihydrotanshinone I and territrem B show peripheral site flexibility.
ACS Med Chem Lett, 4, 2013
|
|
1WU1
 
 | |
4P6L
 
 | |
2XQS
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
1FZV
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PLACENTA GROWTH FACTOR-1 (PLGF-1), AN ANGIOGENIC PROTEIN AT 2.0A RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PLACENTA GROWTH FACTOR | | Authors: | Iyer, S, Leonidas, D.D, Swaminathan, G.J, Maglione, D, Battisti, M, Tucci, M, Persico, M.G, Acharya, K.R. | | Deposit date: | 2000-10-04 | | Release date: | 2001-05-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution.
J.Biol.Chem., 276, 2001
|
|
2XSU
 
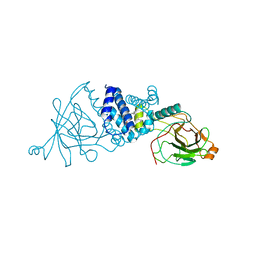 | | Crystal structure of the A72G mutant of Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
3DS1
 
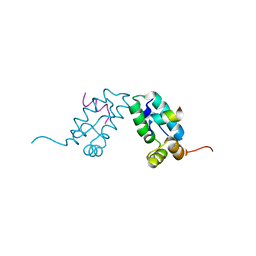 | |
2ORJ
 
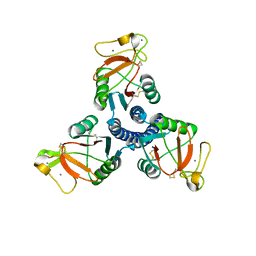 | |
1A48
 
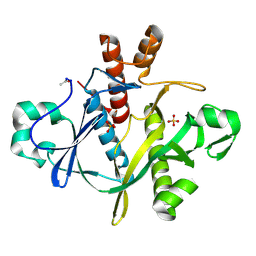 | | SAICAR SYNTHASE | | Descriptor: | PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Levdikov, V.M, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1998-02-12 | | Release date: | 1999-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
Structure, 6, 1998
|
|
3UVF
 
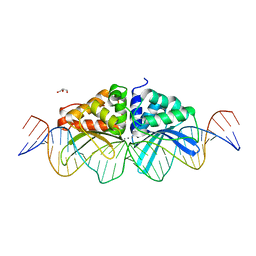 | | Expanding LAGALIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Intron-encoded DNA endonuclease I-HjeMI, ... | | Authors: | Jacoby, K, Metzger, M, Shen, B, Jarjour, J, Stoddard, B, Scharenberg, A. | | Deposit date: | 2011-11-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space.
Nucleic Acids Res., 40, 2012
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
2Y6T
 
 | | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia pestis | | Descriptor: | CHYMOTRYPSINOGEN A, ECOTIN, SULFATE ION | | Authors: | Clark, E.A, Walker, N, Ford, D.C, Cooper, I.A, Oyston, P.C.F, Acharya, K.R. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia Pestis.
J.Biol.Chem., 286, 2011
|
|
2XOK
 
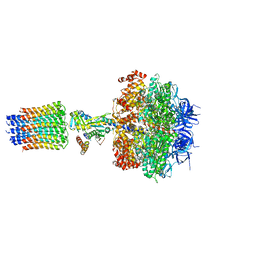 | | Refined structure of yeast F1c10 ATPase complex to 3 A resolution | | Descriptor: | ATP SYNTHASE, ATP SYNTHASE CATALYTIC SECTOR F1 EPSILON SUBUNIT, ATP SYNTHASE SUBUNIT 9, ... | | Authors: | Stock, D, W Leslie, A.G, Walker, J.E. | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular architecture of the rotary motor in ATP synthase.
Science, 286, 1999
|
|
2XQT
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4, DICYCLOHEXYLUREA | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
3DS0
 
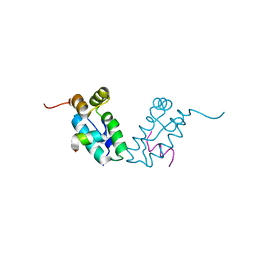 | |
3DS2
 
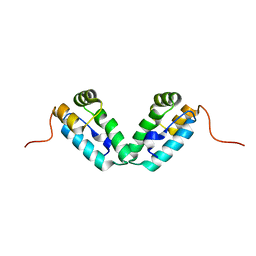 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
1G6D
 
 | | STRUCTURE OF PEPTIDYL-D(CGCAATTGCG) IN THE PRESENCE OF ZINC IONS | | Descriptor: | PEPTIDYL-D(CGCAATTGCG), ZINC ION | | Authors: | Soler-Lopez, M, Malinina, L, Tereshko, V, Zarytova, V, Subirana, J.A. | | Deposit date: | 2000-11-04 | | Release date: | 2002-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction of zinc ions with d(CGCAATTGCG) in a 2.9 A resolution X-ray structure.
J.Biol.Inorg.Chem., 7, 2002
|
|
2YGL
 
 | | The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
4MQR
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with E)-5-hydroxy-4-(((Z)-isonicotinoyldiazenyl)methylene)-6-methyl-1,4-dihydropyridin-3-yl)methyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, [(4Z)-5-hydroxy-6-methyl-4-{[(E)-(pyridin-4-ylcarbonyl)diazenyl]methylidene}-1,4-dihydropyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Transaminase BioA by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
3DSJ
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase Variant (F137L) (At-AOS(F137L), cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOD at 1.60 A Resolution | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
3DS5
 
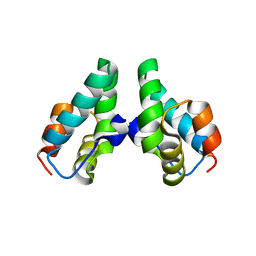 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3TM2
 
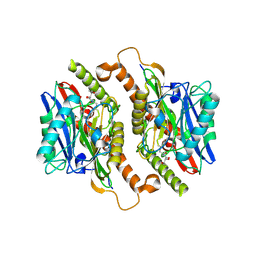 | | Crystal structure of mature ThnT with a covalently bound product mimic | | Descriptor: | (2R)-N-(4-chloro-3-oxobutyl)-2,4-dihydroxy-3,3-dimethylbutanamide, cysteine transferase | | Authors: | Schildbach, J.F, Wright, N.T, Buller, A.R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoproteolytic Activation of ThnT Results in Structural Reorganization Necessary for Substrate Binding and Catalysis.
J.Mol.Biol., 422, 2012
|
|
3DSI
 
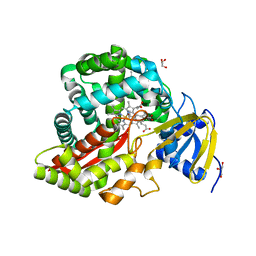 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase (AOS, cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOT at 1.60 A resolution | | Descriptor: | (9Z,11E,13S,15Z)-13-hydroxyoctadeca-9,11,15-trienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
