5RGO
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102248 (Mpro-x0736) | | Descriptor: | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6ERQ
 
 | | Structure of the human mitochondrial transcription initiation complex at the HSP promoter | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
3HLA
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN A2.1 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2.1) (ALPHA CHAIN) | | Authors: | Saper, M.A, Bjorkman, P.J, Wiley, D.C. | | Deposit date: | 1989-10-06 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
J.Mol.Biol., 219, 1991
|
|
3DOL
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with GW695634. | | Descriptor: | N-({4-[({4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}acetyl)amino]-3-methylphenyl}sulfonyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3W1I
 
 | |
1QTP
 
 | |
1QVA
 
 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
4GKN
 
 | | A2-MHC Complex carrying FATGIGIITV | | Descriptor: | Beta-2-microglobulin, FAT Cognate peptide, MHC class I antigen, ... | | Authors: | Sewell, A.K, Rizkallah, P.J, Cole, D.K, Wooldridge, L, Price, D.A. | | Deposit date: | 2012-08-13 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T-cell Receptor-optimized Peptide Skewing of the T-cell Repertoire Can Enhance Antigen Targeting.
J.Biol.Chem., 287, 2012
|
|
2PKP
 
 | | Crystal structure of 3-isopropylmalate dehydratase (leuD)from Methhanocaldococcus Jannaschii DSM2661 (MJ1271) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Homoaconitase small subunit, ZINC ION | | Authors: | Jeyakanthan, J, Gayathri, D.R, Velmurugan, D, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-18 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity determinants of the methanogen homoaconitase enzyme: structure and function of the small subunit
Biochemistry, 49, 2010
|
|
4GM6
 
 | | Crystal structure of PfkB family carbohydrate kinase(TARGET EFI-502146 FROM Listeria grayi DSM 20601 | | Descriptor: | CHLORIDE ION, GLYCEROL, PfkB family carbohydrate kinase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PfkB family carbohydrate kinase FROM Listeria grayi
To be Published
|
|
5R84
 
 | | PanDDA analysis group deposition -- Crystal Structure of COVID-19 main protease in complex with Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4GHC
 
 | | Structure of Y257F variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum at 1.55 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the role of tyrosine 257 of homoprotocatechuate 2,3-dioxygenase in substrate and oxygen activation.
Biochemistry, 51, 2012
|
|
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
1T9S
 
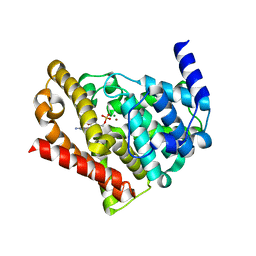 | | Catalytic Domain Of Human Phosphodiesterase 5A in Complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-18 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
1Z6F
 
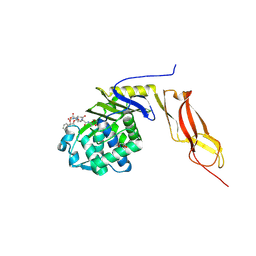 | | Crystal structure of penicillin-binding protein 5 from E. coli in complex with a boronic acid inhibitor | | Descriptor: | GLYCEROL, N1-[(1R)-1-(DIHYDROXYBORYL)ETHYL]-N2-[(TERT-BUTOXYCARBONYL)-D-GAMMA-GLUTAMYL]-N6-[(BENZYLOXY)CARBONYL-L-LYSINAMIDE, Penicillin-binding protein 5 | | Authors: | Nicola, G, Peddi, S, Stefanova, M, Nicholas, R.A, Gutheil, W.G, Davies, C. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli Penicillin-Binding Protein 5 Bound to a Tripeptide Boronic Acid Inhibitor: A Role for Ser-110 in Deacylation.
Biochemistry, 44, 2005
|
|
1TBB
 
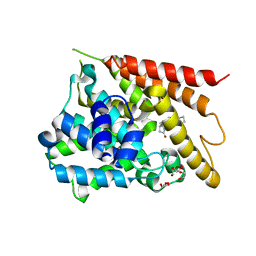 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Rolipram | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ROLIPRAM, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
5RE8
 
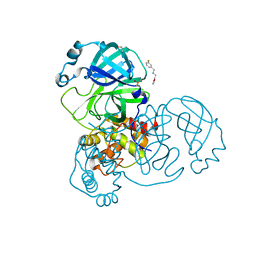 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2737076969 | | Descriptor: | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REN
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102425 | | Descriptor: | 1-[(3R)-3-(1,3-benzothiazol-2-yl)piperidin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1741969146 | | Descriptor: | 1-azanylpropylideneazanium, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102372 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(3S)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-phenylacetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1KCS
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC282 IN COMPLEX WITH PS1 PEPTIDE | | Descriptor: | PC282 IMMUNOGLOBULIN, PS1 peptide | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
5RFU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102121 | | Descriptor: | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5A3D
 
 | | Structural insights into the recognition of cisplatin and AAF-dG lesions by Rad14 (XPA) | | Descriptor: | 5'-D(*DG 5IUP*GP*A 5IUP*GP*AP*CP*G 5IUP*AP*GP*AP*DGP*AP)-3', 5'-D(*DTP*CP*TP*CP*TP*AP*C 8FGP*TP*CP*AP*TP*CP*DAP*CP)-3', DNA REPAIR PROTEIN RAD14, ... | | Authors: | Kuper, J, Koch, S.C, Gasteiger, K.L, Wichlein, N, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1TJE
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase | | Descriptor: | Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1KCU
 
 | | CRYSTAL STRUCTURE OF ANTIBODY PC287 | | Descriptor: | PC287 IMMUNOGLOBULIN | | Authors: | Nair, D.T, Singh, K, Siddiqui, Z, Nayak, B.P, Rao, K.V.S, Salunke, D.M. | | Deposit date: | 2001-11-11 | | Release date: | 2002-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response.
J.Immunol., 168, 2002
|
|
