7G8Y
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1449748885 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(4-methoxyphenyl)glycinamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
2KQP
 
 | |
1QSX
 
 | | SOLUTION NMR STRUCTURE OF THE 2:1 HOECHST 33258-D(CTTTTGCAAAAG)2 COMPLEX | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(CP*TP*TP*TP*TP*GP*CP*AP*AP*AP*AP*G)-3', SODIUM ION | | Authors: | Gavathiotis, E, Sharman, G.J, Searle, M.S. | | Deposit date: | 1999-06-24 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-dependent variation in DNA minor groove width dictates orientational preference of Hoechst 33258 in A-tract recognition: solution NMR structure of the 2:1 complex with d(CTTTTGCAAAAG)(2).
Nucleic Acids Res., 28, 2000
|
|
3ZO1
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | 6-(1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
1CEE
 
 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
5OL3
 
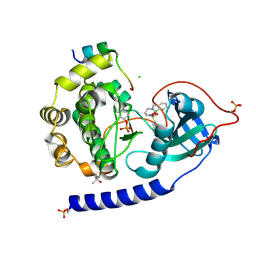 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp013 and RKp117 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
5OK3
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp241 and Fasudil | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, UPF0418 protein FAM164A, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Diamondoid Amino Acid-Based Peptide Kinase A Inhibitor Analogues.
Chemmedchem, 14, 2019
|
|
3ZDT
 
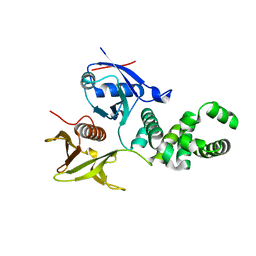 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7QG0
 
 | | Inhibitor-induced hSARM1 duplex | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zalk, R, Kahzma, T, Guez-Haddad, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A duplex structure of SARM1 octamers stabilized by a new inhibitor.
Cell.Mol.Life Sci., 80, 2022
|
|
3ZJY
 
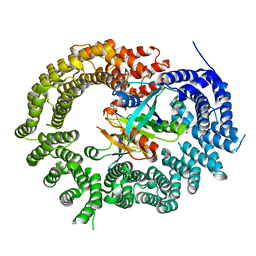 | | Crystal Structure of Importin 13 - RanGTP - eIF1A complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 1A, X-CHROMOSOMAL, GTP-BINDING NUCLEAR PROTEIN RAN, ... | | Authors: | Gruenwald, M, Lazzaretti, D, Bono, F. | | Deposit date: | 2013-01-21 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis for the Nuclear Export Activity of Importin13.
Embo J., 32, 2013
|
|
3ZO3
 
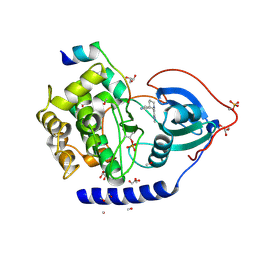 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | 6-(2,9-DIAZASPIRO[5.5]UNDECAN-2-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
5OYX
 
 | |
5OZ7
 
 | |
5OZI
 
 | |
5OZT
 
 | |
5P06
 
 | |
5OYU
 
 | |
5OZ3
 
 | |
7QUI
 
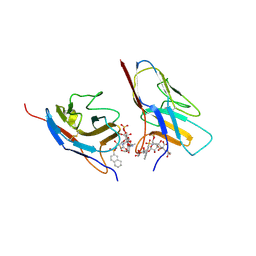 | | Crystal structure of the N-terminal domain of Siglec-8 in complex with sulfonamide sialoside analogue | | Descriptor: | (2~{S},4~{S},5~{R},6~{R})-5-acetamido-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-acetamido-2-(hydroxymethyl)-4,6-bis(oxidanyl)oxan-3-yl]oxy-3,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-4-yl]oxy-6-[(1~{R},2~{R})-3-(naphthalen-2-ylsulfonylamino)-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Oyenarte, I, Atxabal, U, Nycholat, C, Franconetti, A, Quintana, J.I, Delgado, S, Unione, L, Paulson, J, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
5OYZ
 
 | |
5OZ9
 
 | |
7QU6
 
 | | Crystal structure of the N-terminal domain of Siglec-8 | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Atxabal, U, Nycholat, C.M, Oyenarte, I, Paulson, J.C, Franconetti, A, Quintana, J.I, Unione, L, Delgado, S, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
5OZL
 
 | |
5OZW
 
 | |
5P09
 
 | |
