4IX7
 
 | |
4IT4
 
 | | Crystal structure of residues 1-211 of CG17282 | | Descriptor: | ACETIC ACID, CG17282, FORMIC ACID, ... | | Authors: | Agyekum, B, Bouyain, S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Noncanonical FK506-Binding Protein BDBT Binds DBT to Enhance Its Circadian Function and Forms Foci at Night.
Neuron, 80, 2013
|
|
4X8W
 
 | | dsRBD3 of Loquacious | | Descriptor: | GLYCEROL, Loquacious, isoform B | | Authors: | Jakob, L, Treiber, N, Treiber, T, Stotz, M, Meister, G. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Structure of the dsRBD3 of Loquacious reveals features for dimerization and Dicer1-interaction
To Be Published
|
|
7B3Y
 
 | | Structure of a nanoparticle for a COVID-19 vaccine candidate | | Descriptor: | Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses.
Nat Commun, 12, 2021
|
|
3PC3
 
 | |
3GMJ
 
 | | Crystal structure of MAD MH2 domain | | Descriptor: | Protein mothers against dpp | | Authors: | Wu, J.W, Wang, C. | | Deposit date: | 2009-03-14 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MH2 domain of Drosophila Mad
SCI.CHINA, SER.C: LIFE SCI., 52, 2009
|
|
5GWM
 
 | |
4YL8
 
 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
5FYX
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD16 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, FREQUENIN 2, ... | | Authors: | Martinez-Gonzalez, L, Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4PJJ
 
 | | MYOSIN VI (MD-INSERT2-CAM, DELTA-INSERT1) post-rigor state - long soaking with PO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
5A7D
 
 | | Tetrameric assembly of LGN with Inscuteable | | Descriptor: | INSCUTEABLE, PINS | | Authors: | Culurgioni, S, Mari, S, Bonetto, G, Gallini, S, Brennich, M, Round, A, Mapelli, M. | | Deposit date: | 2015-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of the Insc:Lgn Tetramer Reveals a New Function of Lgn in Promoting Asymmetric Cell Divisions
To be Published
|
|
4XDA
 
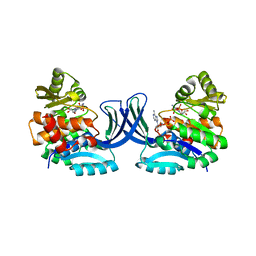 | | Vibrio cholerae O395 Ribokinase complexed with Ribose, ADP and Sodium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribokinase, SODIUM ION, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation
Adv.Exp.Med.Biol., 842, 2015
|
|
3MVD
 
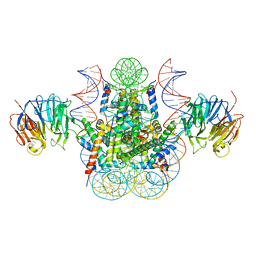 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
4X9H
 
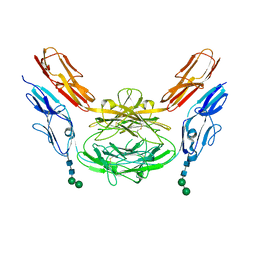 | | Crystal structure of Dscam1 isoform 8.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform AP, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4RWT
 
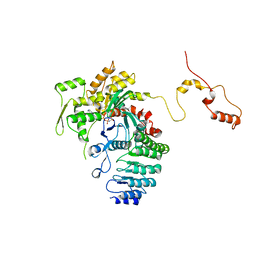 | | Structure of actin-Lmod complex | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3EL2
 
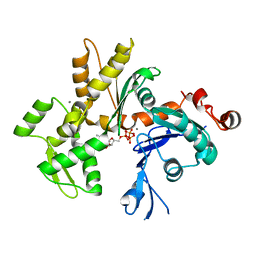 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
4XHQ
 
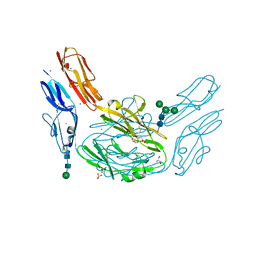 | |
4QUC
 
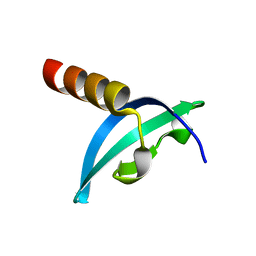 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4XID
 
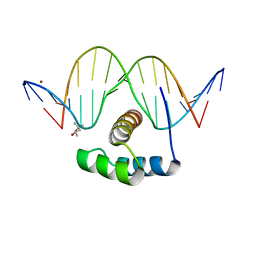 | | AntpHD with 15bp DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
3EKS
 
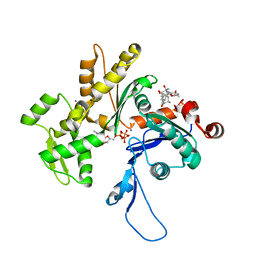 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
5HNY
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2016-11-02 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
4RKG
 
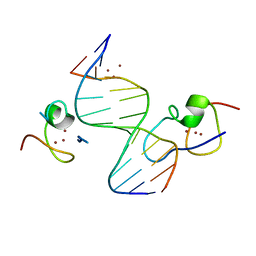 | | Structure of the MSL2 CXC domain bound with a non-specific (GC)6 DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), E3 ubiquitin-protein ligase msl-2, ZINC ION | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
4RV0
 
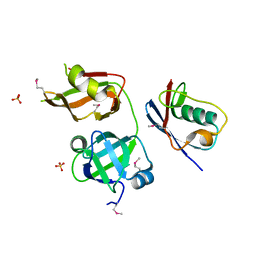 | | Crystal structure of TN complex | | Descriptor: | Nuclear protein localization protein 4 homolog, SULFATE ION, Transitional endoplasmic reticulum ATPase TER94 | | Authors: | Hao, Q, Jiao, S, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2014-11-23 | | Release date: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of TN complex
To be Published
|
|
5ABX
 
 | |
5ABY
 
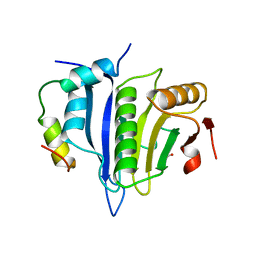 | | Complex of C. elegans eIF4E-3 with the 4E-binding protein Mextli | | Descriptor: | 4E-BINDING PROTEIN MEXTLI, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-3, GLYCEROL, ... | | Authors: | Peter, D, Weichenrieder, O. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mextli Proteins Use Both Canonical Bipartite and Novel Tripartite Binding Modes to Form Eif4E Complexes that Display Differential Sensitivity to 4E-BP Regulation
Genes Dev., 29, 2015
|
|
