8C41
 
 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*GP*AP*AP*T)-3'), ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8C3W
 
 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8C3U
 
 | | Crystal Structure of human IL-1beta in complex with a low molecular weight antagonist | | Descriptor: | (S)-4'-hydroxy-3'-(6-methyl-2-oxo-3-(1H-pyrazol-4-yl)indolin-3-yl)-[1,1'-biphenyl]-2,4-dicarboxylic acid, Interleukin-1 beta | | Authors: | Rondeau, J.-M, Lehmann, S, Koch, E. | | Deposit date: | 2022-12-28 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Discovery of a selective and biologically active low-molecular weight antagonist of human interleukin-1 beta.
Nat Commun, 14, 2023
|
|
8C3P
 
 | |
8C3O
 
 | | Crystal structure of autotaxin gamma and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3N
 
 | |
8C3J
 
 | | Stapled peptide SP2 in complex with humanised RadA mutant HumRadA22 | | Descriptor: | 2-[(4,6-diethyl-1,3,5-triazin-2-yl)-methyl-amino]ethanoic acid, Breast cancer type 2 susceptibility protein, DNA repair and recombination protein RadA | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | A recombinant approach for stapled peptide discovery yields inhibitors of the RAD51 recombinase.
Chem Sci, 14, 2023
|
|
8C3I
 
 | | Dark state of PAS-GAF fragment from Deinococcus radiodurans phytochrome | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Madan Kumar, S, Sebastian, W. | | Deposit date: | 2022-12-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dark state of PAS-GAF fragment from Deinococcus radiodurans phytochrome
To Be Published
|
|
8C3F
 
 | | Double mutant I(L177)H/F(M197)H structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2022-12-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H.
Membranes (Basel), 13, 2023
|
|
8C3E
 
 | | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein) | | Descriptor: | Engineered protein LCB2, GLYCEROL | | Authors: | Korban, S.A, Mikhailovskii, O.V, Luzik, D.A, Gurzhiy, V.V, Levkina, A.D, Kharkov, B.B, Skrynnikov, N.R. | | Deposit date: | 2022-12-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein)
To Be Published
|
|
8C3B
 
 | | X-ray structure of RNase A upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | (1,3-dimethyl-2~{H}-imidazol-2-yl)-oxidanyl-oxidanylidene-ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tetrakis(oxidanyl)ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tris(oxidanyl)ruthenium, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Ruthenium(II)-arene Complexes with Glycosylated Carbene Ligands: Synthesis, Characterization, Antiproliferative Activity, In Solution and Crystallographic Evidences of Macromolecule Binding
To Be Published
|
|
8C2S
 
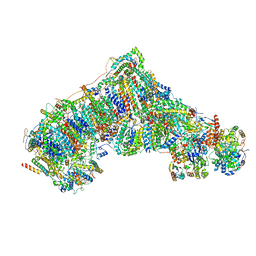 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 1). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8C2F
 
 | |
8C2E
 
 | |
8C28
 
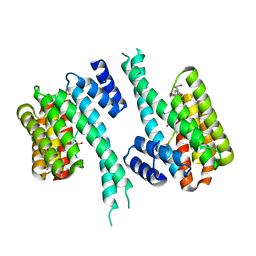 | | 14-3-3 in complex with PyrinpS208pS242 | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyrin | | Authors: | Lau, R, Ottmann, C, Hann, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 14-3-3/Pyrin complex
To Be Published
|
|
8C26
 
 | |
8C25
 
 | | purine nucleoside phosphorylase in complex with JS-375 | | Descriptor: | CHLORIDE ION, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8C24
 
 | |
8C23
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (monoclinic form M200T#m) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8C1Y
 
 | | Small molecule stabilizer for 14-3-3/ChREBP (Cmd 30) | | Descriptor: | 14-3-3 protein sigma, Carbohydrate-responsive element-binding protein, [2-[2-[[2,2-bis(fluoranyl)-2-phenyl-ethyl]amino]-2-oxidanylidene-ethoxy]phenyl]phosphonic acid | | Authors: | Pennings, M.A.M, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.
Biorxiv, 2024
|
|
8C1X
 
 | |
8C1V
 
 | | SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sb92, ... | | Authors: | Huiskonen, J.T, Rissanen, I, Hannula, L. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Intranasal trimeric sherpabody inhibits SARS-CoV-2 including recent immunoevasive Omicron subvariants.
Nat Commun, 14, 2023
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1T
 
 | |
