4N35
 
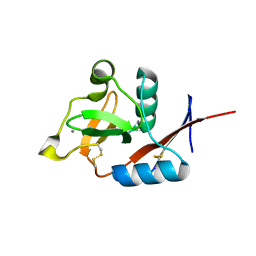 | | Structure of langerin CRD I313 complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, C-type lectin domain family 4 member K, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
5VCW
 
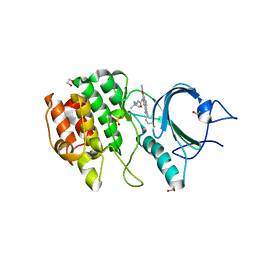 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
6KZF
 
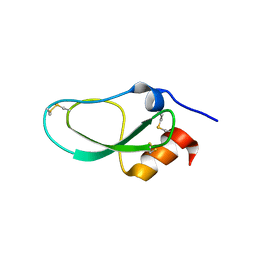 | | Racemic X-ray Structure of Calcicludine | | Descriptor: | D-calcicludine, Kunitz-type serine protease inhibitor homolog calcicludine | | Authors: | Qu, Q, Gao, S, Liu, L. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Synthesis of Disulfide Surrogate Peptides Incorporating Large-Span Surrogate Bridges Through a Native-Chemical-Ligation-Assisted Diaminodiacid Strategy
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8GDS
 
 | |
5VEU
 
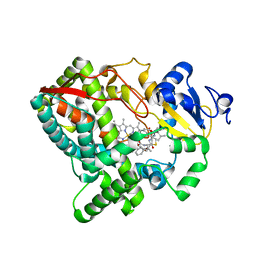 | | Human Cytochrome P450 3A5 (CYP3A5) | | Descriptor: | Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE, RITONAVIR | | Authors: | Hsu, M.-H, Johnson, E.F. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The X-Ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5.
Mol. Pharmacol., 93, 2018
|
|
7UCX
 
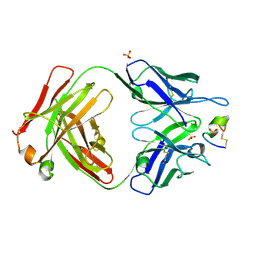 | | LRP8 11H1 Fab complexed to a cyclized CR1 peptide | | Descriptor: | 11H1 Fab Heavy chain, 11H1 Fab Light chain, Cyclized CR1 peptide, ... | | Authors: | Argiriadi, M.A, Deng, K, Egan, D, Gao, L, Gizatullin, F, Harlan, J, Karaoglu, D, Qiu, W, Goodearl, A. | | Deposit date: | 2022-03-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The use of cyclic peptide antigens to generate LRP8 specific antibodies
Front Drug Discov (Lausanne), 2, 2023
|
|
7D6L
 
 | |
5VCZ
 
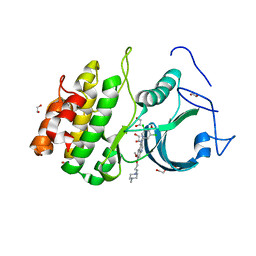 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Bosutinib isomer | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VD7
 
 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH RAC-IV-098, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
6A4O
 
 | | HEWL crystals soaked in 2.5M GuHCl for 20 minutes | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANIDINE, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
6L17
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
5VH2
 
 | |
5VEE
 
 | | PAK4 kinase domain in complex with FRAX486 | | Descriptor: | 6-(2,4-dichlorophenyl)-8-ethyl-2-{[3-fluoro-4-(piperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhang, E.Y, Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PAK4 crystal structures suggest unusual kinase conformational movements.
Biochim. Biophys. Acta, 1866, 2018
|
|
5VEI
 
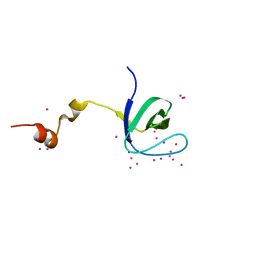 | | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2 | | Descriptor: | Sorbin and SH3 domain-containing protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Huang, H, Gu, J, Liu, K, Sidhu, S.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2
To be Published
|
|
4NN9
 
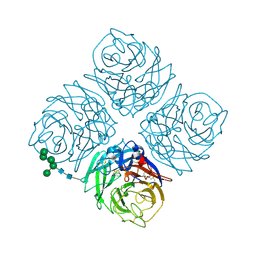 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
8GSQ
 
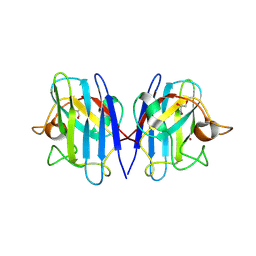 | | Structure based studies reveal an atypical antipsychotic drug candidate - Paliperidone as a potent hSOD1 modulator with implications in ALS treatment. | | Descriptor: | (9~{R})-3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-9-oxidanyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, GLYCEROL, Superoxide dismutase [Cu-Zn], ... | | Authors: | Aouti, S, Padmanabhan, B. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of an antipsychotic drug, paliperidone, as a modulator of human superoxide dismutase 1: a potential therapeutic target in amyotrophic lateral sclerosis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6E93
 
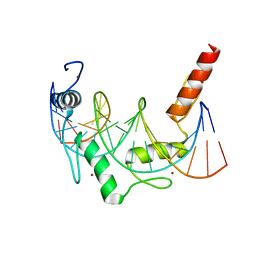 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
7M9W
 
 | | HIV-1 Protease (I84V) in Complex with NR02-73 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with NR02-73
To Be Published
|
|
7M9S
 
 | | HIV-1 Protease WT (NL4-3) in Complex with NR01-141 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with NR01-141
To Be Published
|
|
5VHB
 
 | |
4NEB
 
 | | Previously de-ionized HEW lysozyme batch crystallized in 0.5 M MnCl2 | | Descriptor: | CHLORIDE ION, Lysozyme C, MANGANESE (II) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-10-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7M9X
 
 | | HIV-1 Protease (I84V) in Complex with NR02-79 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{(2-ethylbutyl)[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with NR02-79
To Be Published
|
|
5VIB
 
 | |
7M9V
 
 | | HIV-1 Protease (I84V) in Complex with NR01-141 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with NR01-141
To Be Published
|
|
7M9T
 
 | | HIV-1 Protease WT (NL4-3) in Complex with NR02-73 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with NR02-73
To Be Published
|
|
