1A9R
 
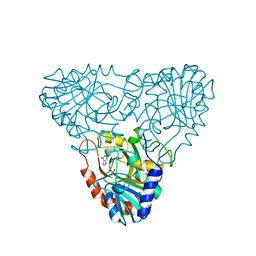 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH HYPOXANTHINE AND SULFATE | | Descriptor: | HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A2X
 
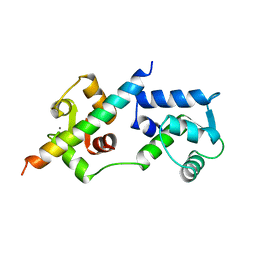 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A9P
 
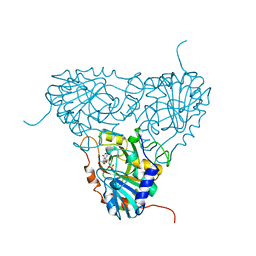 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH 9-DEAZAINOSINE AND PHOSPHATE | | Descriptor: | 9-DEAZAINOSINE, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A3K
 
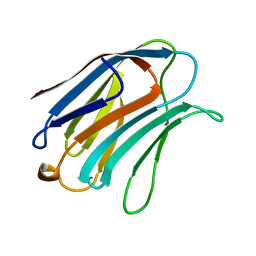 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
1CWF
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 2-VAL CYCLOSPORIN | | Descriptor: | CYCLOSPORIN D, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1A8G
 
 | | HIV-1 PROTEASE IN COMPLEX WITH SDZ283-910 | | Descriptor: | HIV-1 PROTEASE, benzyl [(1R)-1-({(1S,2S,3S)-1-benzyl-2-hydroxy-4-({(1S)-1-[(2-hydroxy-4-methoxybenzyl)carbamoyl]-2-methylpropyl}amino)-3-[(4-methoxybenzyl)amino]-4-oxobutyl}carbamoyl)-2,2-dimethylpropyl]carbamate | | Authors: | Kallen, J, Billich, A, Scholz, D, Auer, M, Kungl, A. | | Deposit date: | 1998-03-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and conformational dynamics of the HIV-1 protease in complex with the inhibitor SDZ283-910: agreement of time-resolved spectroscopy and molecular dynamics simulations.
J.Mol.Biol., 286, 1999
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1A2Z
 
 | |
1CWH
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 3-D-SER CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-07 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1JFR
 
 | |
1BA2
 
 | |
1CWM
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 4 MEILE CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-26 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1CWL
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 4 4-HYDROXY-MELEU CYCLOSPORIN | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-26 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
372D
 
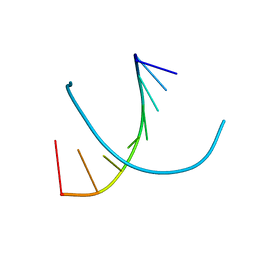 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
412D
 
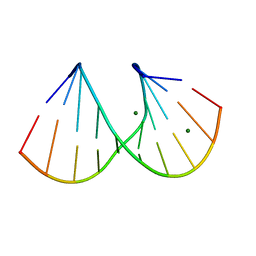 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHYL-[TRI(OXYETHYL)] RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(126)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
410D
 
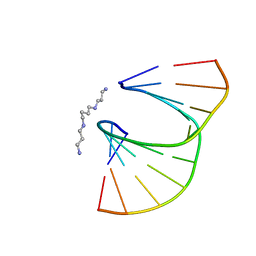 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-ETHOXYMETHYLENE RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T38)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
411D
 
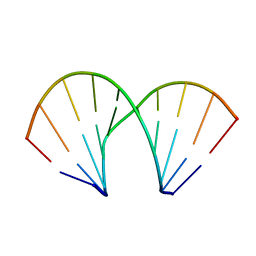 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHOXYETHYL RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T39)P*AP*CP*GP*C)-3') | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
1BKW
 
 | | p-Hydroxybenzoate hydroxylase (phbh) mutant with cys116 replaced by ser (c116s) and arg44 replaced by lys (r44k), in complex with fad and 4-hydroxybenzoic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H, Schreuder, H.A, Van Berkel, W.J. | | Deposit date: | 1998-07-13 | | Release date: | 1998-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
Eur.J.Biochem., 231, 1995
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AP1
 
 | |
1C11
 
 | | INTERCALATED D(TCCCGTTTCCA) DIMER, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*GP*TP*TP*TP*CP*CP*A)-3') | | Authors: | Gallego, J, Chou, S.H, Reid, B.R. | | Deposit date: | 1998-07-15 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer.
J.Mol.Biol., 273, 1997
|
|
1GB4
 
 | |
2IGD
 
 | |
1A3P
 
 | | ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | EPIDERMAL GROWTH FACTOR | | Authors: | Barnham, K, Torres, A, Alewood, D, Alewood, P, Domagala, T, Nice, E, Norton, R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-29 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Role of the 6-20 disulfide bridge in the structure and activity of epidermal growth factor.
Protein Sci., 7, 1998
|
|
