5QYF
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry F02a | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, R-1,2-PROPANEDIOL, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYG
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry F08a | | Descriptor: | 4-[(methylamino)methyl]phenol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYK
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry H12a | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, R-1,2-PROPANEDIOL, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYH
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry G04a | | Descriptor: | A1 cistron-splicing factor AAR2, N-(4-methoxyphenyl)acetamide, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYJ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry H11a | | Descriptor: | A1 cistron-splicing factor AAR2, N-cyclopentyl-N'-{[(2R)-oxolan-2-yl]methyl}urea, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1YED
 
 | |
1YEC
 
 | |
1P17
 
 | | Hypoxanthine Phosphoribosyltransferase from Trypanosoma cruzi, K68R mutant, complexed with the product IMP | | Descriptor: | INOSINIC ACID, hypoxanthine phosphoribosyltransferase | | Authors: | Medrano, F.J, Eakin, A.E, Craig III, S.P. | | Deposit date: | 2003-04-11 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions at the dimer interface influence the relative efficiencies for purine nucleotide synthesis and pyrophosphorolysis in a phosphoribosyltransferase.
J.Mol.Biol., 335, 2004
|
|
1P18
 
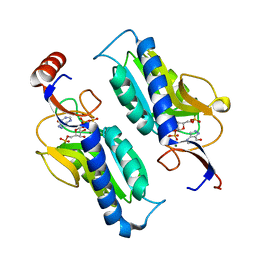 | | Hypoxanthine Phosphoribosyltransferase from Trypanosoma cruzi, K68R mutant, ternary substrates complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, MAGNESIUM ION, ... | | Authors: | Canyuk, B, Eakin, A.E, Craig III, S.P. | | Deposit date: | 2003-04-11 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions at the dimer interface influence the relative efficiencies for purine nucleotide synthesis and pyrophosphorolysis in a phosphoribosyltransferase
J.Mol.Biol., 335, 2004
|
|
1P2U
 
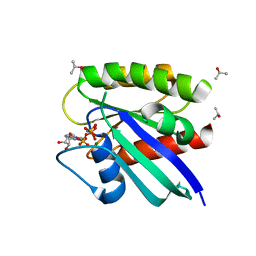 | | H-Ras in 50% isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Buhrman, G.K, de Serrano, V, Mattos, C. | | Deposit date: | 2003-04-16 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Organic solvents order the dynamic switch II in Ras crystals
Structure, 11, 2003
|
|
2CMK
 
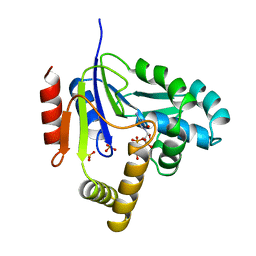 | | CYTIDINE MONOPHOSPHATE KINASE IN COMPLEX WITH CYTIDINE-DI-PHOSPHATE | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Briozzo, P. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
3KS0
 
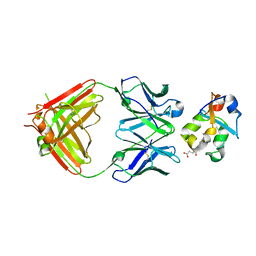 | | Crystal structure of the heme domain of flavocytochrome b2 in complex with Fab B2B4 | | Descriptor: | Cytochrome b2, mitochondrial, Fragment Antigen Binding B2B4, ... | | Authors: | Golinelli-Pimpaneau, B, Lederer, F, Le, K.H.D. | | Deposit date: | 2009-11-20 | | Release date: | 2010-05-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the functional importance of the heme domain mobility in flavocytochrome b2.
J.Mol.Biol., 400, 2010
|
|
2EWG
 
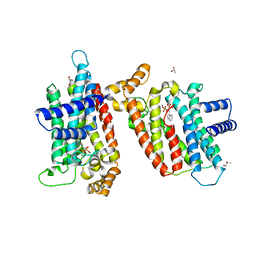 | | T. brucei Farnesyl Diphosphate Synthase Complexed with Minodronate | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Cao, R, Mao, J, Gao, Y, Robinson, H, Odeh, S, Goddard, A, Oldfield, E. | | Deposit date: | 2005-11-03 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Solid-state NMR, crystallographic, and computational investigation of bisphosphonates and farnesyl diphosphate synthase-bisphosphonate complexes.
J.Am.Chem.Soc., 128, 2006
|
|
1YEE
 
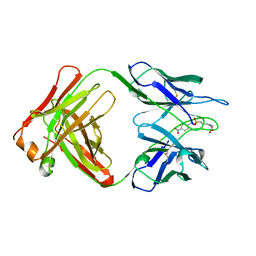 | |
5RPA
 
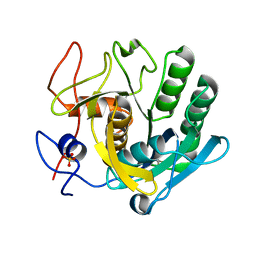 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen E3a | | Descriptor: | 3-[(4E)-4-imino-5,6-dimethylfuro[2,3-d]pyrimidin-3(4H)-yl]-N,N-dimethylpropan-1-amine, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3QPO
 
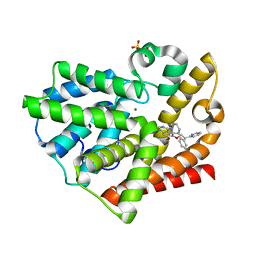 | | Structure of PDE10-inhibitor complex | | Descriptor: | 7-methoxy-4-[(3S)-3-phenylpiperidin-1-yl]-6-[2-(pyridin-2-yl)ethoxy]quinazoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2011-02-14 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
3QPN
 
 | | Structure of PDE10-inhibitor complex | | Descriptor: | 6-methoxy-7-[2-(quinolin-2-yl)ethoxy]quinazoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2011-02-14 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
3ESF
 
 | |
4DK4
 
 | | Crystal Structure of Trypanosoma brucei dUTPase with dUpNp, Ca2+ and Na+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, CALCIUM ION, Deoxyuridine triphosphatase, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-03 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
4DKB
 
 | | Crystal Structure of Trypanosoma brucei dUTPase with dUpNp and Ca2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, CALCIUM ION, Deoxyuridine triphosphatase | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-03 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
7FKP
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04E08 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, propyl (2S)-2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]propanoate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLI
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05D09 from the F2X-Universal Library | | Descriptor: | (2S)-2-[(pyridin-2-yl)sulfanyl]propanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLF
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05C06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl 3-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)propanoate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7M1E
 
 | |
7FO1
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07G03 from the F2X-Universal Library | | Descriptor: | 3-[(2-methylpropyl)sulfanyl]-N-(pyridin-3-yl)propanamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
