3UGO
 
 | |
3QLP
 
 | | X-ray structure of the complex between human alpha thrombin and a modified thrombin binding aptamer (mTBA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-02-03 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thrombin-aptamer recognition: a revealed ambiguity.
Nucleic Acids Res., 39, 2011
|
|
3UGP
 
 | |
7QXS
 
 | | Cryo-EM structure of human telomerase-DNA-TPP1-POT1 complex (with POT1 side chains) | | Descriptor: | Adrenocortical dysplasia homolog (Mouse), isoform CRA_a, Histone H2A, ... | | Authors: | Sekne, Z, Ghanim, G.E, van Roon, A.M.M, Nguyen, T.H.D. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of human telomerase recruitment by TPP1-POT1.
Science, 375, 2022
|
|
7QXB
 
 | | Cryo-EM map of human telomerase-DNA-TPP1-POT1 complex (sharpened map) | | Descriptor: | Adrenocortical dysplasia homolog (Mouse), isoform CRA_a, Histone H2A, ... | | Authors: | Sekne, Z, Ghanim, G.E, van Roon, A.M.M, Nguyen, T.H.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of human telomerase recruitment by TPP1-POT1.
Science, 375, 2022
|
|
7P0B
 
 | |
7P0M
 
 | |
7P09
 
 | |
7OXO
 
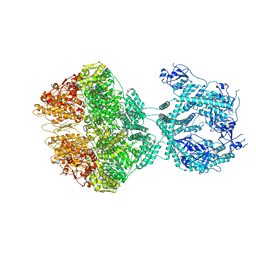 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
6V9D
 
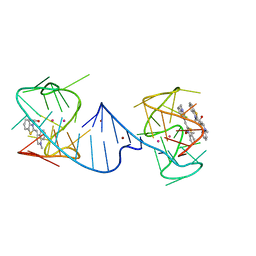 | |
6V9B
 
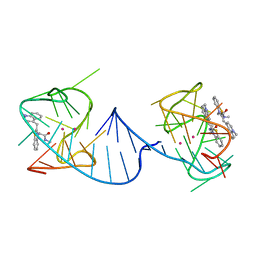 | |
7KVU
 
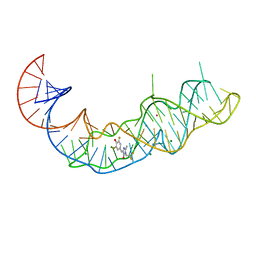 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVT
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T with iridium (III) ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVV
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, Squash RNA aptamer bound to DFHO | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KPF
 
 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
7A8R
 
 | | Structure of RecQL from Bos taurus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase, MAGNESIUM ION, ... | | Authors: | Rety, S, Chen, W.F, Xi, X.G. | | Deposit date: | 2020-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Endogenous Bos taurus RECQL is predominantly monomeric and more active than oligomers.
Cell Rep, 36, 2021
|
|
5UC6
 
 | |
5W0Z
 
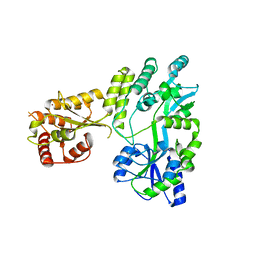 | |
1DX1
 
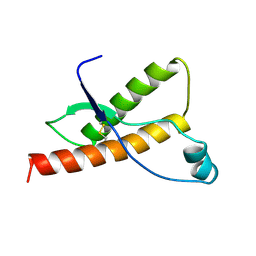 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6FWS
 
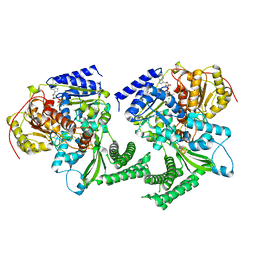 | | Structure of DinG in complex with ssDNA and ADPBeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DinG, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Cheng, K, Wigley, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
5W0U
 
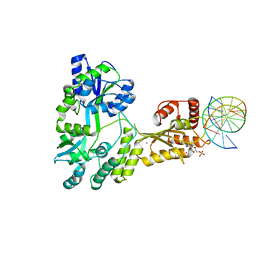 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6FWR
 
 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
1DX0
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EG6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D(CG(5-BRU)ACG) COMPLEXES TO A PHENAZINE | | Descriptor: | 5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3', 9-BROMO-PHENAZINE-1-CARBOXYLIC ACID (2-DIMETHYLAMINO-ETHYL)-AMIDE, BROMIDE ION, ... | | Authors: | Cardin, C.J, Denny, W.A, Hobbs, J.R, Thorpe, J.H. | | Deposit date: | 2000-02-14 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions.
Biochemistry, 39, 2000
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
