2WSA
 
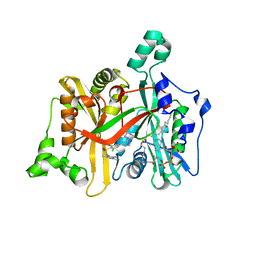 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
9QRS
 
 | | Crystal structure of CERK6 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, J.X.J, Gysel, K, Stougaard, J, Andersen, K.R. | | Deposit date: | 2025-04-04 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
3NRB
 
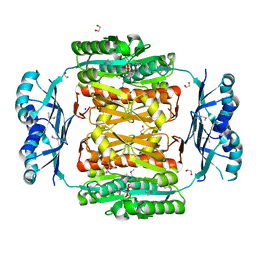 | |
2WXL
 
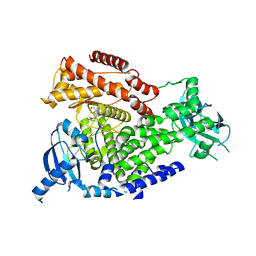 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with ZSTK474. | | Descriptor: | 2-(difluoromethyl)-1-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-1H-benzimidazole, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
4GRH
 
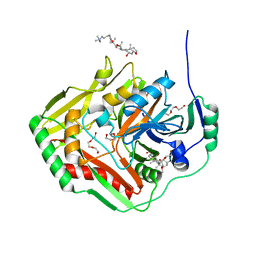 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
3GVB
 
 | |
9H9E
 
 | | Cryo-EM structure of the human GABAA receptor alpha1 subunit in complex with the assembly factor NACHO/TMEM35A | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Hooda, Y, Sente, A, Judy, R.M, Smalinskaite, L, Peak-Chew, S, Naydenova, K, Malinauskas, T, Hardwick, S.W, Chirgadze, D.Y, Aricescu, A.R, Hegde, R.S. | | Deposit date: | 2024-10-30 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of NACHO-mediated assembly of pentameric ligand-gated ion channels.
Biorxiv, 2024
|
|
4GSY
 
 | |
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
4BZC
 
 | | Crystal structure of the tetrameric dGTP-bound wild type SAMHD1 catalytic core | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION, ... | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
3O2T
 
 | |
1FM8
 
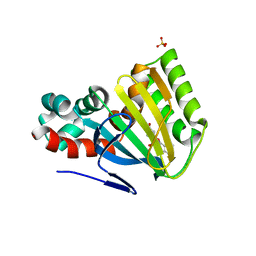 | | CHALCONE ISOMERASE COMPLEXED WITH 5,4'-DIDEOXYFLAVANONE | | Descriptor: | 7-HYDROXY-2-PHENYL-CHROMAN-4-ONE, CHALCONE-FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Noel, J.P. | | Deposit date: | 2000-08-16 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences.
J.Biol.Chem., 277, 2002
|
|
9KWH
 
 | | Crystal structure of S. aureus tryptophanyl-tRNA synthetase complexed with chuangxinmycin | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, Tryptophan--tRNA ligase | | Authors: | Ren, Y, Qiao, H, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2024-12-05 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
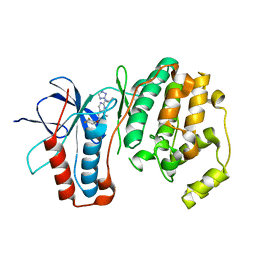
3FMH
 
 | |
1RHA
 
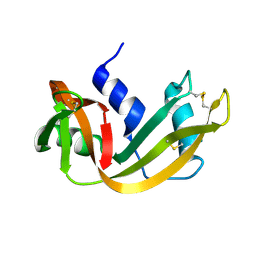 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2O1I
 
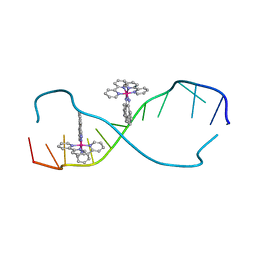 | | RH(BPY)2CHRYSI complexed to mismatched DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3', bis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)[chrysene-5,6-diiminato(2-)-kappa~2~N,N']rhodium(4+) | | Authors: | Pierre, V.C, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2006-11-28 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B84
 
 | | Crystal structure of the human BTB domain of the Krueppel related Zinc Finger Protein 3 (HKR3) | | Descriptor: | 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, Zinc finger and BTB domain-containing protein 48 | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, T, Salah, E, Pilka, E, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the Human BTB domain of the Krueppel related Zinc Finger Protein 3 (HKR3).
To be Published
|
|
9KXB
 
 | | Crystal structure of S. aureus tryptophanyl-tRNA synthetase complexed with 3-methylchuangxinmycin | | Descriptor: | (6~{R})-5,5-dimethyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, Tryptophan--tRNA ligase | | Authors: | Ren, Y, Qiao, H, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2024-12-06 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
2KGP
 
 | | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by Novantrone (Mitoxantrone) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, RNA (25-MER) | | Authors: | Zheng, S, Chen, Y, Donahue, C.P, Wolfe, M.S, Varani, G. | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Chem.Biol., 16, 2009
|
|
3UWI
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
1XXL
 
 | |
9KWW
 
 | | Crystal structure of S. aureus tryptophanyl-tRNA synthetase complexed with 3-demethylchuangxinmycin | | Descriptor: | (6~{R})-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(11),3,8(12),9-tetraene-6-carboxylic acid, Tryptophan--tRNA ligase | | Authors: | Ren, Y, Qiao, H, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2024-12-06 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
7XBB
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 23a | | Descriptor: | (2~{R},4~{S})-6-ethyl-2-[(2~{E})-2-hydroxyiminoethyl]-2,8-dimethyl-4-(2-methylprop-1-enyl)-3,4-dihydropyrano[3,2-c][1,8]naphthyridin-5-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.100009 Å) | | Cite: | Structure-based optimization of Toddacoumalone as highly potent and selective PDE4 inhibitors with anti-inflammatory effects.
Biochem Pharmacol, 202, 2022
|
|
3H51
 
 | |
1KTU
 
 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
