3A9J
 
 | | Crystal structure of the mouse TAB2-NZF in complex with Lys63-linked di-ubiquitin | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2, Ubiquitin, ZINC ION | | Authors: | Sato, Y, Yoshikawa, A, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3
Embo J., 28, 2009
|
|
8TRG
 
 | |
3CQL
 
 | | Crystal Structure of GH family 19 chitinase from Carica papaya | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endochitinase, ... | | Authors: | Huet, J, Rucktoa, P, Clantin, B, Azarkan, M, Looze, Y, Villeret, V, Wintjens, R. | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray Structure of Papaya Chitinase Reveals the Substrate Binding Mode of Glycosyl Hydrolase Family 19 Chitinases.
Biochemistry, 47, 2008
|
|
1PJ4
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ2
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
4D4O
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P64 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
1NAA
 
 | | Cellobiose Dehydrogenase Flavoprotein Fragment in Complex with Cellobionolactam | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Henriksson, G, Pettersson, G, Vasella, A, Divne, C. | | Deposit date: | 2002-11-27 | | Release date: | 2003-01-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of the reductive half-reaction in cellobiose dehydrogenase
J.BIOL.CHEM., 278, 2003
|
|
4DDT
 
 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
7E40
 
 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | Authors: | Zhou, J, Hu, Q, Yao, D, Xing, W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
3A1Q
 
 | | Crystal structure of the mouse RAP80 UIMs in complex with Lys63-linked di-ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin interaction motif-containing protein 1 | | Authors: | Sato, Y, Yoshikawa, A, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80
Embo J., 28, 2009
|
|
4DUW
 
 | | E. coli (lacZ) beta-galactosidase (G974A) in complex with allolactose | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Lo, S, Janzcewicz, L.J, Dugdale, M.L, Huber, R.E. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Explanation for Allolactose (lac operon inducer) Synthesis by lacZ
beta-Galactosidase and the Evolutionary Relationship between Allolactose
synthesis and the lac Repressor
J.Biol.Chem., 288, 2013
|
|
1XGP
 
 | |
2INP
 
 | | Structure of the Phenol Hydroxylase-Regulatory Protein Complex | | Descriptor: | FE (III) ION, Phenol hydroxylase component phL, Phenol hydroxylase component phM, ... | | Authors: | Sazinsky, M.S, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
3E0O
 
 | | Crystal structure of MsrB | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Park, A.K, Shin, Y.J, Kim, Y.K, Chi, Y.M, Hwang, K.Y. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Kinetic Analysis of an MsrA-MsrB Fusion Protein from Streptococcus pneumoniae
Mol.Microbiol., 72, 2009
|
|
3ODK
 
 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4D4P
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P65 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
1PJ3
 
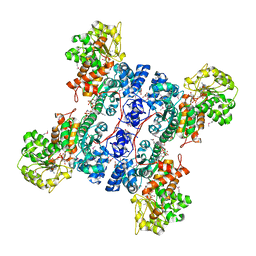 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
7N2V
 
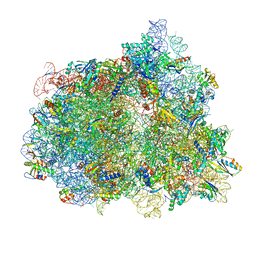 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7EF9
 
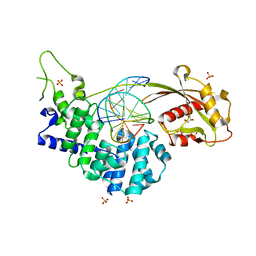 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EF8
 
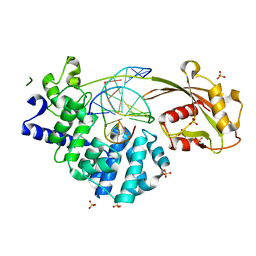 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EFA
 
 | |
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2ZR6
 
 | |
1XLU
 
 | | X-Ray Structure Of Di-Isopropyl-Phosphoro-Fluoridate (Dfp) Inhibited Butyrylcholinesterase after Aging | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Asojo, O.A, Borgstahl, G.E.O, Masson, P, Lockridge, O. | | Deposit date: | 2004-09-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Role of Water in Aging of Human Butyrylcholinesterase Inhibited by Echothiophate: The Crystal Structure Suggests Two Alternative Mechanisms of Aging
Biochemistry, 44, 2005
|
|
