5Q1W
 
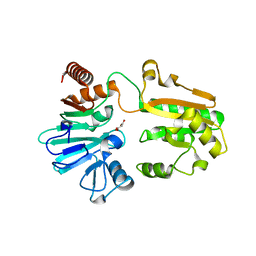 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000546a | | Descriptor: | 1~{H}-benzimidazol-2-ylcyanamide, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3A79
 
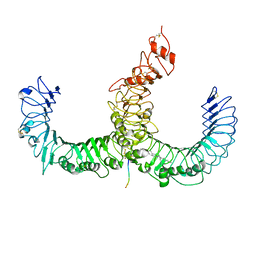 | | Crystal structure of TLR2-TLR6-Pam2CSK4 complex | | Descriptor: | (2S)-propane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kang, J.Y, Jin, M.S, Lee, J.-O. | | Deposit date: | 2009-09-20 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer
Immunity, 31, 2009
|
|
7Y2B
 
 | |
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4Q0I
 
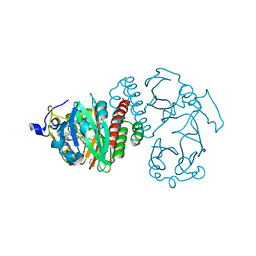 | | Deinococcus radiodurans BphP PAS-GAF D207A mutant | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ISOPROPYL ALCOHOL | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
6ADS
 
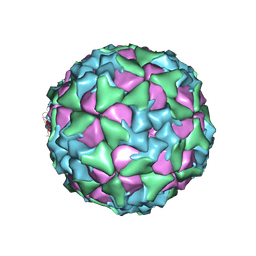 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5SQ0
 
 | |
5SPZ
 
 | |
3FLI
 
 | | Discovery of XL335, a Highly Potent, Selective and Orally-Active Agonist of the Farnesoid X Receptor (FXR) | | Descriptor: | 1-methylethyl 3-[(3,4-difluorophenyl)carbonyl]-1,1-dimethyl-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Bile acid receptor | | Authors: | Foster, P.G, Stout, T.J. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of XL335 (WAY-362450), a highly potent, selective, and orally active agonist of the farnesoid X receptor (FXR).
J.Med.Chem., 52, 2009
|
|
5SPX
 
 | |
3UGF
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
5SQ2
 
 | |
5SRJ
 
 | |
2WDC
 
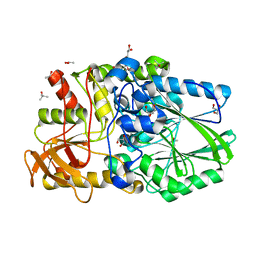 | | Termus thermophilus Sulfate thiohydrolase SoxB in complex with glycerol | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Sauve, V, Roversi, P, Leath, K.J, Garman, E.F, Antrobus, R, Lea, S.M, Berks, B.C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the Hydrolysis of a Sulfur-Sulfur Bond Based on the Crystal Structure of the Thiosulfohydrolase Soxb.
J.Biol.Chem., 284, 2009
|
|
5SQ4
 
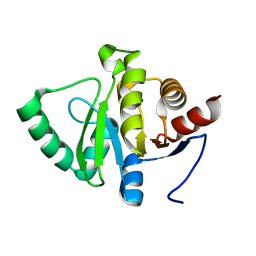 | |
3UGH
 
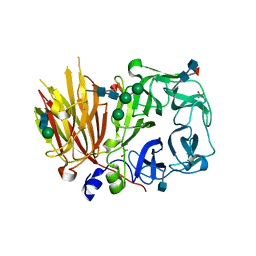 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
9M78
 
 | | Consensus map of UCC118 Rool RNA at 2.26-angstrom resolution | | Descriptor: | UCC118 Rool RNA | | Authors: | Zhang, S, Yi, R, An, L, Li, S, Zhang, K. | | Deposit date: | 2025-03-10 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structural insights into higher-order natural RNA-only multimers.
Nat.Struct.Mol.Biol., 32, 2025
|
|
2A7C
 
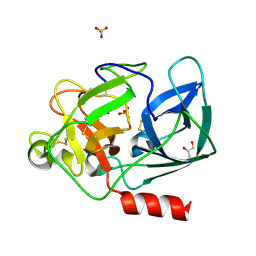 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | Elastase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5SQ8
 
 | |
5SQB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894390 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894388 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4XQN
 
 | | Crystal structure of AgrA LytTR domain in complex with promoters | | Descriptor: | 1,2-ETHANEDIOL, Accessory gene regulator A, DNA (5'-D(*AP*CP*AP*GP*TP*TP*AP*AP*GP*AP*AP*T)-3'), ... | | Authors: | Gopal, B, Rajasree, K. | | Deposit date: | 2015-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Biochem Biophys Rep, 6, 2016
|
|
3IMX
 
 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
5SQ5
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894407 - (R,S) and (S,S) isomers | | Descriptor: | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4B73
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
