5SBK
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z1258992717 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBO
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z2856434878 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,4-dimethoxyphenyl)methyl]morpholine, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBP
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z1229798311 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-6-fluorobenzoic acid, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.275 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
3I1U
 
 | | Carboxypeptidase A Inhibited by a Thiirane Mechanism-Based inactivator | | Descriptor: | (2S,3R)-2-benzyl-3-sulfanylbutanoic acid, Carboxypeptidase A1 (Pancreatic), GLYCEROL, ... | | Authors: | Fernandez, D. | | Deposit date: | 2009-06-28 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | The X-ray structure of carboxypeptidase A inhibited by a thiirane mechanism-based inhibitor
Chem.Biol.Drug Des., 75, 2010
|
|
3I2J
 
 | | Cocaine Esterase, wild type, without a ligand | | Descriptor: | CHLORIDE ION, Cocaine esterase, GLYCEROL, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
5SBX
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z53825479 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
1NIV
 
 | |
7HLI
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z2017861827 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S,4S)-1,2,3,4-tetrahydro-1,4-(epiminomethano)naphthalen-10-yl]ethan-1-one, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3I4X
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus in complex with Trp and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XFD
 
 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
4HZX
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
2A70
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
3A1J
 
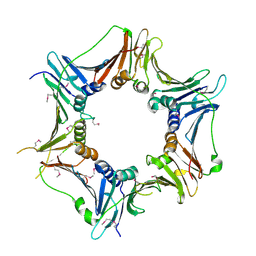 | | Crystal structure of the human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Sohn, S.Y, Cho, Y. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp
J.Mol.Biol., 390, 2009
|
|
5PAI
 
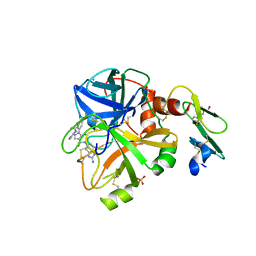 | | human factor VIIa in complex with N-(2-amino-1H-benzimidazol-5-yl)-1-[3-[[(3,5-dimethyl-1,2-oxazol-4-yl)carbamoylamino]methyl]phenyl]-5-hydroxypyrazole-4-carboxamide at 1.73A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII heavy chain, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
3HVJ
 
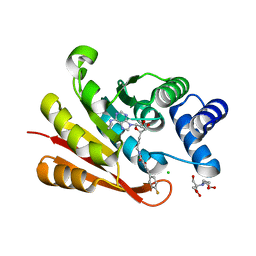 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-propyladenine-containing bisubstrate inhibitor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5BOD
 
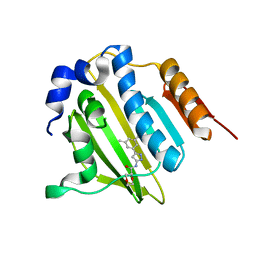 | | Crystal structure of Streptococcus pneumonia ParE inhibitor | | Descriptor: | (2R)-N-[3-(3,5-dimethylphenyl)-1H-pyrazol-5-yl]-1,4-dioxane-2-carboxamide, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
to be published
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
4I1E
 
 | |
3HXV
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
3HZU
 
 | |
4QDG
 
 | |
2XRU
 
 | | AURORA-A T288E COMPLEXED WITH PHA-828300 | | Descriptor: | 3-({[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]CARBONYL}AMINO)-N-[(1R)-1-PHENYLPROPYL]-1H-THIENO[3,2-C]PYRAZOLE-5-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Bindi, S, Fancelli, D, Alli, C, Berta, D, Bertrand, J.A, Cameron, A.D, Cappella, P, Carpinelli, P, Cervi, G, Croci, W, D'Anello, M, Forte, B, LauraGiorgini, M, Marsiglio, A, Moll, J, Pesenti, E, Pittala, V, Pulici, M, Riccardi-Sirtori, F, Roletto, F, Soncini, C, Storici, P, Varasi, M, Volpi, D, Zugnoni, P, Vianello, P. | | Deposit date: | 2010-09-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bioorg.Med.Chem., 18, 2010
|
|
5POB
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with E13683b | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-1,3-dimethyl-1,3-dihydro-2H-benzimidazol-2-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5SMC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2033637875 | | Descriptor: | N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
