3HY0
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
5BS6
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, transcriptional regulator AraR | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
4POB
 
 | |
5XMW
 
 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3HWW
 
 | |
5RXN
 
 | |
5S8M
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N11511a (space group C2) | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | XChem group deposition
To Be Published
|
|
2XL3
 
 | | WDR5 IN COMPLEX WITH AN RBBP5 PEPTIDE AND HISTONE H3 PEPTIDE | | Descriptor: | GLYCEROL, HISTONE H3.1, RETINOBLASTOMA-BINDING PROTEIN 5, ... | | Authors: | Odho, Z, Southall, S.M, Wilson, J.R. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterisation of a Novel Wdr5 Binding Site that Recruits Rbbp5 Through a Conserved Motif and Enhances Methylation of H3K4 by Mll1.
J.Biol.Chem., 285, 2010
|
|
6T2P
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16 | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
3I4W
 
 | | Crystal Structure of the third PDZ domain of PSD-95 | | Descriptor: | ACETATE ION, Disks large homolog 4 | | Authors: | Camara-Artigas, A, Gavira, J.A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4A X-ray structures
J.Struct.Biol., 170, 2010
|
|
3I60
 
 | |
5BQN
 
 | | Crystal structure of the LHn fragment of botulinum neurotoxin type D, mutant H233Y E230Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Botulinum neurotoxin type D,Botulinum neurotoxin type D | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
5S8G
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N00804d (space group C2) | | Descriptor: | 1,3-benzothiazole-6-carboxylic acid, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
4QC3
 
 | | Crystal structure of human BAZ2B bromodomain in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, diacetylated histone 4 peptide (H4K8acK12ac) | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4C1Q
 
 | | Crystal structure of the PRDM9 SET domain in complex with H3K4me2 and AdoHcy. | | Descriptor: | GLYCEROL, HISTONE H3.1, HISTONE-LYSINE N-METHYLTRANSFERASE PRDM9, ... | | Authors: | Mathioudakis, N, Cusack, S, Kadlec, J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of Prdm9.
Cell Rep., 5, 2013
|
|
4I4E
 
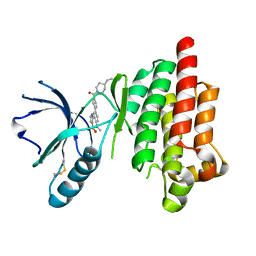 | | Structure of Focal Adhesion Kinase catalytic domain in complex with hinge binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, [4-(2-hydroxyethyl)piperidin-1-yl][4-(5-methyl-4,4-dioxido-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-yl)phenyl]methanone | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3HVI
 
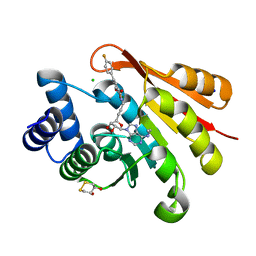 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-ethyladenine-containing bisubstrate inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4PXL
 
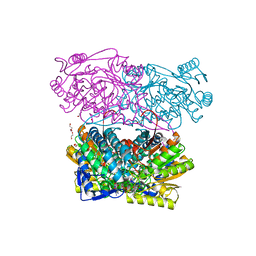 | | Structure of Zm ALDH2-3 (RF2C) in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytosolic aldehyde dehydrogenase RF2C, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
3HXU
 
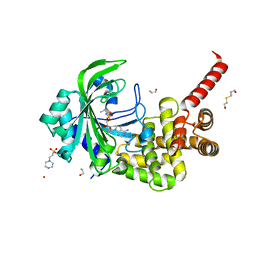 | | Crystal Structure of catalytic fragment of E. coli AlaRS in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
1NBP
 
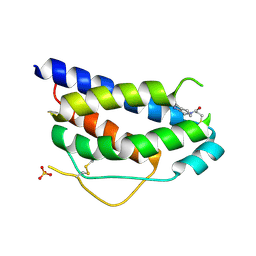 | | Crystal Structure Of Human Interleukin-2 Y31C Covalently Modified At C31 With 3-Mercapto-1-(1,3,4,9-tetrahydro-B-carbolin-2-yl)-propan-1-one | | Descriptor: | 3-MERCAPTO-1-(1,3,4,9-TETRAHYDRO-B-CARBOLIN-2-YL)-PROPAN-1-ONE, Interleukin-2, SULFATE ION | | Authors: | Hyde, J, Braisted, A.C, Randal, M, Arkin, M.R. | | Deposit date: | 2002-12-03 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of cooperative ligand binding in the adaptive region of interleukin-2
Biochemistry, 42, 2003
|
|
5SBN
 
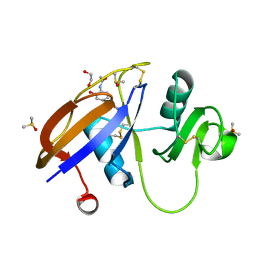 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z57040482 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.181 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
6DQ3
 
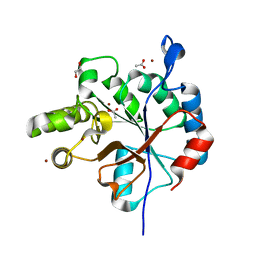 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
5SBL
 
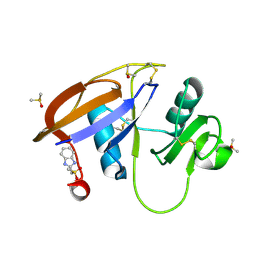 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z126932614 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
3FCN
 
 | |
